Abstract
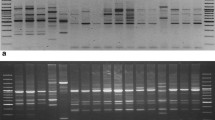
Nepenthes khasiana Hook. f. belonging to monotypic family Nepenthaceae is a rare, endangered, dioecious member of the carnivorous plant found in North-East India. The plant is endemic to the Indian state of Meghalaya and is distributed throughout the state from West Khasi hills to East Khasi hills, Jaintia hills and East, West to South Garo hills from 1,000 to ca. 1,500 m altitude. Multi-locus analysis using PCR based Random Amplified Polymorphic DNA (RAPD) and Inter Simple Sequence Repeats (ISSR) markers were used for the first time to assess the genetic diversities of N. khasiana Hook. f. collected from different parts of Meghalaya. It was observed that RAPD analysis showed more polymorphism than ISSR fingerprinting in revealing genetic polymorphism in N. khasiana Hook. f. The result of cluster analysis by using UPGMA method showed that the groups based on pooled RAPD–ISSR genetic similarity were more similar than the groups based on RAPD. Furthermore, genetic similarity reveals variability within the population at Jarain of Jaintia hills, while between populations the Baghmara region differs from the others with at least 40% dissimilarity. The results show a broad range of genetic diversity within the populations of N. khasiana Hook. f.





Similar content being viewed by others
References
Avise JC (1994) Mitochondrial DNA polymorphism and a connection between genetics and demography of relevance to conservation. Conserv Biol 9:686–690. doi:10.1046/j.1523-1739.1995.09030686.x
Bramwell D (1972) In: Valentine DH (ed) Endemism in the flora of the Canary Islands. Taxonomy, phytogeography and evolution. Academic Press, London, pp 141–159
Cannon JR, Lojanapiwanta V, Raston CL, Sinchai W, White AH (1980) The quinones of Nepenthes rafflesiana. Aust J Chem 33:1075–1093
Chaveerach A, Tanomtang A, Sudmood R, Tanee T (2006) Genetic diversity among geographically distributed population of Nepenthes mirabilis. Biologia (Bratisl) 61:295–298. doi:10.2478/s11756-006-0054-4
Doyle JJ, Doyle JL (1990) Isolation of plant DNA from fresh tissue. Focus 12:13–15
Eguiarte LE, Pinero D (1990) Genética de la conservación: leones vemos, genes no sabemos. Ciencias 4:34–47
Esselman EJ, Jianqiang L, Crawford DJ, Windus JL, Wolfe AD (1999) Clonal diversity in the rare Calamagrostis porteri ssp. insperata (Poaceae): comparative results for allozymes and random amplified polymorphic DNA (RAPD) and intersimple sequence repeat (ISSR) markers. Mol Ecol 8:443–451. doi:10.1046/j.1365-294X.1999.00585.x
Fang DQ, Roose ML (1997) Identification of closely related citrus cultivars with inter-simple sequence repeat markers. Theor Appl Genet 95:408–417. doi:10.1007/s001220050577
Fracaro F, Zacaria J, Echeverrigaray S (2005) RAPD based genetic relationships between populations of three chemotypes of Cunila galoioides Benth. Biochem Syst Ecol 33:409–417. doi:10.1016/j.bse.2004.10.017
Frankel HO, Soulé ME (1981) Conservation and evolution Cambridge. Cambridge University Press, Cambridge
Gallie DR, Chang SC (1997) Signal transduction in the carnivorous plants Sarracenia purpurea. Plant Physiol 115:1461–1471. doi:10.1104/pp.115.4.1461
Gary AJ (1987) Genetic change during succession in plants. In: Gary AJ, Edwards PJ, McCauley J (eds) Colonization, succession and stability. Blackwell Scientifc Publications, Oxford
Gitzendanner MA, Soltis PS (2000) Patterns of genetic variation in rare and widespread plant congeners. Am J Bot 87:783–792. doi:10.2307/2656886
Givnish TJ, Burkhardt EL, Happel RE, Weintraub JW (1984) Carnivory in the bromeliad Brocchinia reducta, with a cost/benefit model for the general restriction of carnivorous plants to sunny, moist, nutrient poor habitats. Am Nat 124:479–497. doi:10.1086/284289
González-Astorga J, Castillo-Campos G (2004) Genetic variability of narrow endemic tree Antirhea aromatica Castilo-Campos and Lorence (Rubiaceae, Guettardeae) in a tropical forest of Mexico. Ann Bot (Lond) 93:521–528. doi:10.1093/aob/mch070
González-Astorga J, Cruz-Angón A, Flores-Palacios A, Vovides AP (2004) Diversity and genetic structure of the Mexican endemic epiphyte: Tillandsia achyrostachys E. Morr. ex Baker var. achyrostachys (Bromeliaceae). Ann Bot (Lond) 94:545–551. doi:10.1093/aob/mch171
Harbone JB (1982) Introduction to ecological biochemistry. Academic Press, New York
Heubl G, Bringmann G, Meimberg H (2006) Molecular phylogeny and character evolution of carnivorous plant families in caryophyllales. Plant Biol 8:821–830. doi:10.1055/s-2006-924460
Hoebee SE, Young AG (2001) Low neighbourhood size and high interpopulation differentiation in the endangered shrub Grevillea iaspicula McGill (Proteaceae). Heredity 86:489–496. doi:10.1046/j.1365-2540.2001.00857.x
Holsinger KE, Gottlieb LD (1991) Conservation of rare and endangered plants: principles and prospects. In: Falk DA, Holsinger KE (eds) Genetics and conservation of rare plants. Oxford University Press, New York, pp 195–208
Hooker JD (1886) The flora of British India. vol 5. pp 68–71
Jain SK, Baishya AK (1977) Nepenthes khasiana: an endangered species. Hornbill: 17–18
Jain SK, Sastri ARK (1980) Threatened plants of India- a state-of-the-art report Bot Survy India, Calcutta 48
Jayaram K, Prasad MNV (2005) Rapidly in vitro multiplied Drosera as reliable source for plumbagin bioprospection. Curr Sci 89:447–448
Joseph J, Joseph K (1986) Insectivorous plants of khasi and Jaintia hills, Meghalaya, India. Botanical Survey of India, Calcutta
Kanjilal UN, Kanjilal PC, De RN, Das A (1940) Nepentheses. Flora of Assam, vol IV, pp 25
Kharkongar P, Joseph J (1981) Folklore medico botany of rural Khasi and Jantia tribes in Meghalaya. In: Jain SK (ed) Glimpses of Indian Ethnobotany. Oxford IBH Publishing Co., New Delhi, pp 124–136
Khoshbakht K, Hammer K (2007) Threatened and rare ornamental plants. J Agric Rural Dev Trop Subtrop 108:19–39
Kumar YK, Haridasan S, Rao RR (1980) Ethnobotanical notes on certain medicinal plants among some Garo people around Balphakram Sanctury in Meghalaya. Bull Bot Surv India 22:161–165
Kurata K, Jaffré T, Setoguchi H (2008) Genetic diversity and geographical structure of the pitcher plant Nepenthes vieillardii in New Caledonia: a chloroplast DNA haplotype analysis. Am J Bot 95:1632–1644. doi:10.3732/ajb.0800129
Lewontin RC, Hubby JL (1966) A molecular approach to the study of genetic heterozygosity in natural populations. II. Amount of variation and degree of heterozygosity in natural populations of Drosophila pseudoobscura. Genetics 54:595–609
Li JM, Jin ZX (2006) High genetic differentiation revealed by RAPD analysis of narrowly endemic Sinocalycanthus chinensis Cheng et S. Y. Chang, an endangered species of China. Biochem Syst Ecol 34:725–735. doi:10.1016/j.bse.2006.06.010
Lim SH, Phua DCY, Tan HTW (2000) Primer design and optimization for RAPD analysis of Nepenthes. Biol Plant 43:153–155. doi:10.1023/A:1026535920714
Loveless MD, Hamrick JL (1984) Ecological determinants of genetic structure in plant population. Annu Rev Ecol Syst 15:65–95. doi:10.1146/annurev.es.15.110184.000433
Maki M, Asada Y (1998) High genetic variability revealed by allozymic loci in the narrow endemic fern Polystichum otomasui (Dryoperidaceae). Heredity 80:604–610. doi:10.1046/j.1365-2540.1998.00328.x
Mantel NA (1967) The detection of disease clustering and a generalized regression approach. Cancer Res 27:209–220
Mao AA, Kharbuli P (2002) Distribution and status of Nepenthes khasiana Hook. f.—a rare endemic pitcher plant of Meghalaya, India. Phytotaxonomy 2:77–83
Mokkamul P, Chaveerach A, Sudmoon R, Tavee T (2007) Species identification and sex determination of the genus Nepenthes (Nepenthaceae). Pak J Biol Sci 10:561–567. doi:10.3923/pjbs.2007.561.567
Mukerjee A, Dam DP, Dam N (1984) Pitcher plant—an ornamental climber of Meghalaya. Ind Hort April–June 1:6–18
Myers N, Mittermeier RA, Mittermeier CG, da Fonseca GAB, Kent J (2000) Biodiversity hotspots for conservation priorities. Nature 403:853–858
Oostermeijer JGB, Luijten SH, denNijs JCM (2003) Integrating demographic and genetic approaches in plant conservation. Biol Conserv 113:389–398. doi:10.1016/S0006-3207(03)00127-7
Prevost A, Wilkinson MJ (1999) A new system of comparing PCR primers applied to ISSR fingerprinting of potato cultivars. Theor Appl Genet 98:107–112. doi:10.1007/s001220051046
Rao TA, Shanware PG, Tribedi GN (1969) A note on the pitcher plant habitat in Assam. Ind For 95:611–613
Reynolds EH (1987) Early treatment and prognosis in epilepsy. Epilepsia 28:97–106. doi:10.1111/j.1528-1157.1987.tb03633.x
Sokal RR, Sneath PHA (1963) Principles of Numeric Taxonomy. Freeman, San Francisco, p 359
Soltis PS, Soltis DE (1991) Multiple origins of the allotetraploid Tragopogon mirus (Compositae): rDNA evidence. Syst Bot 16:407–413. doi:10.2307/2419333
Tamaki I, Seteku S, Tomaru N (2008) Genetic variation and differentiation in populations of a threatened tree, Magnolia stellata: factors influencing the level of within-population genetic variation. Heredity 100:415–423. doi:10.1038/sj.hdy.6801097
Thomson RH (1987) Naturally occurring quinones III. Chapman & Hall, New York
Williams CG, Hamrick JL (1996) Elite populations for conifer breeding and gene conservation. Can J Res 26:453–461. doi:10.1139/x26-051
Young YA, Boyel T, Brown T (1996) The population genetic consequences of habitat fragmentation for plants. Trends Ecol Evol 11:413–418. doi:10.1016/0169-5347(96)10045-8
Zietkiewicz E, Rafalski A, Labuda D (1994) Genome fingerprinting by simple sequence repeat (SSR)-anchored polymerase chain reaction amplification. Genomics 20:176–183. doi:10.1006/geno.1994.1151
Acknowledgment
Authors are grateful to Dr. P. G. Rao, Director NEIST for providing facilities and constant encouragement to undertake this work.
Author information
Authors and Affiliations
Corresponding author
Additional information
B. S. Bhau and K. Medhi contributed equally.
Rights and permissions
About this article
Cite this article
Bhau, B.S., Medhi, K., Sarkar, T. et al. PCR based molecular characterization of Nepenthes khasiana Hook. f.—pitcher plant. Genet Resour Crop Evol 56, 1183–1193 (2009). https://doi.org/10.1007/s10722-009-9444-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10722-009-9444-0