Abstract
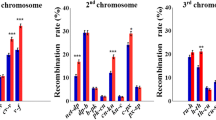
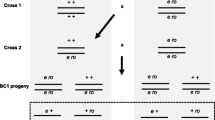
Meiotic recombination is evolutionarily ambiguous, as being associated with both benefits and costs to its bearers, with the resultant dependent on a variety of conditions. While existing theoretical models explain the emergence and maintenance of recombination, some of its essential features remain underexplored. Here we focus on one such feature, recombination plasticity, and test whether recombination response to stress is fitness-dependent. We compare desiccation stress effects on recombination rate and crossover interference in chromosome 3 between desiccation-sensitive and desiccation-tolerant Drosophila lines. We show that relative to desiccation-tolerant genotypes, desiccation-sensitive genotypes exhibit a significant segment-specific increase in single- and double-crossover frequencies across the pericentromeric region of chromosome 3. Significant changes (relaxation) in crossover interference were found for the interval pairs flanking the centromere and extending to the left arm of the chromosome. These results indicate that desiccation is a recombinogenic factor and that desiccation-induced changes in both recombination rate and crossover interference are fitness-dependent, with a tendency of less fitted individuals to produce more variable progeny. Such dependence may play an important role in the regulation of genetic variation in populations experiencing environmental challenges.


Similar content being viewed by others

References
Aggarwal DD, Rashkovetsky E, Michalak P et al (2015) Experimental evolution of recombination and crossover interference in Drosophila caused by directional selection for stress-related traits. BMC Biol 13:101. https://doi.org/10.1186/s12915-015-0206-5
Agrawal AF, Hadany L, Otto SP (2005) The evolution of plastic recombination. Genetics 171:803–812. https://doi.org/10.1534/genetics.105.041301
Baker BS, Hall JC (1976) Meiotic mutants: genie control of meiotic recombination and chromosome segregation. In: Ashburner M, Novitski E (eds) The genetics and biology of Drosophila. Academic Press, New York, pp 351–434
Bomblies K, Higgins JD, Yant L (2015) Meiosis evolves: adaptation to external and internal environments. New Phytol 208:306–323. https://doi.org/10.1111/nph.13499
Bomblies K, Jones G, Franklin C, Zickler D, Kleckner N (2016) The challenge of evolving stable polyploidy: could an increase in “crossover interference distance” play a central role? Chromosoma 125(2):287–300
Borodin PM (1987) Stress and genetic variation. Genetika 23:1003–1010 (in Russian)
Borodin PM, Belyaev DK (1980a) Effect of stress on the rate of crossing-over in the 2nd chromosome of domestic mouse. Dokl Akad Nauk SSSR 253:727–729 (in Russian)
Borodin PM, Belyaev DK (1980b) Effect of emotional stress on recombination rate in chromosome 1 of house mouse. Dokl Akad Nauk SSSR 286:726–728 (in Rissian)
Bownes M, Scott A, Shirras A (1988) Dietary components modulate yolk protein gene transcription in Drosophila melanogaster. Development 103:119–128
Chandley AC (1968) The effect of X-rays on female germ cells of Drosophila melanogaster. III. A comparison with heat-treatment on crossing-over in the X-chromosome. Mutat Res 5:93–107. https://doi.org/10.1017/cbo9781107415324.004
Charlesworth B (1993) Directional selection and the evolution of sex and recombination. Genet Res 61:205–224. https://doi.org/10.1017/s0016672300031372
Davring L, Sunner M (1973) Female meiosis and embryonic mitosis in Drosophila melanogaster. Hereditas 73:51–64. https://doi.org/10.1111/j.1601-5223.1973.tb01067.x
Denell RE, Keppy DO (1979) The nature of genetic recombination near the third chromosome centromere of Drosophila melanogaster. Genetics 93:117–130
Fagegaltier D, König A, Gordon A et al (2014) A genome-wide survey of sexually dimorphic expression of Drosophila miRNAs identifies the steroid hormone-induced miRNA let-7 as a regulator of sexual identity. Genetics 198:647–668. https://doi.org/10.1534/genetics.114.169268
Foss E, Lande R, Stahl FW, Steinberg CM (1993) Chiasma interference as a function of genetic distance. Genetics 133:681–691
Fujitani Y, Mori S, Kobayashi I (2002) A reaction-diffusion model for interference in meiotic crossing over. Genetics 161:365–372
Gessler DDG, Xu S (2000) Meiosis and the evolution of recombination at low mutation rates. Genetics 156:449–456
Goldstein DB, Bergman A, Feldman MW (1993) The evolution of interference: reduction of recombination among three loci. Theor Popul Biol 44:246–259
Gorlov IP, Borodin PM (1986) Effect of emotional stress on the frequency of meiotic abnormalities in male mice. Genetika 22:1019–1024 (in Russian)
Graubard MA (1932) Inversion in Drosophila melanogaster. Genetics 17:81–105
Graubard MA (1934) Temperature effect on interference and crossing over. Genetics 19:83–94
Grell RF (1978) A comparison of heat and interchromosomal effects on recombination and interference in Drosophila melanogaster. Genetics 89:65–77
Gueijman A, Ayali A, Ram Y, Hadany L (2013) Dispersing away from bad genotypes: the evolution of fitness-associated dispersal (FAD) in homogeneous environments. BMC Evol Biol 13:125. https://doi.org/10.1186/1471-2148-13-125
Hadany L, Beker T (2003a) On the evolutionary advantage of fitness-associated recombination. Genetics 165:2167–2179
Hadany L, Beker T (2003b) Fitness-associated recombination on rugged adaptive landscapes. J Evol Biol 16:862–870. https://doi.org/10.1046/j.1420-9101.2003.00586.x
Hadany L, Otto SP (2007) The evolution of condition-dependent sex in the face of high costs. Genetics 176:1713–1727. https://doi.org/10.1534/genetics.107.074203
Hayman DL, Parsons PA (1960) The effect of temperature, age and an inversion on recombination values and interference in the X-chromosome of Drosophila melanogaster. Genetics 32:74–88
Hingston PA, Piercey MJ, Truelstrup Hansen L (2015) Genes associated with desiccation and osmotic stress in Listeria monocytogenes as revealed by insertional mutagenesis. Appl Environ Microbiol 81:5350–5362. https://doi.org/10.1128/aem.01134-15
Hiraizumi Y (1971) Spontaneous recombination in Drosophila melanogaster males. Proc Natl Acad Sci USA 68:268–270. https://doi.org/10.1073/pnas.68.2.268
Hoffmann AA, Parsons PA (1991) Evolutionary genetics and environmental stress. Oxford University Press, New York
Huang DW, Sherman BT, Zheng X et al (2009) Extracting biological meaning from large gene lists with DAVID. Curr Protoc Bioinform 27:13.11.1–13.11.13. https://doi.org/10.1002/0471250953.bi1311s27
Hunter CM, Huang W, Mackay TFC, Singh ND (2016) The genetic architecture of natural variation in recombination rate in Drosophila melanogaster. PLoS Genet 12:e1005951. https://doi.org/10.1371/journal.pgen.1005951
Jackson S, Nielsen DM, Singh ND (2015) Increased exposure to acute thermal stress is associated with a non-linear increase in recombination frequency and an independent linear decrease in fitness in Drosophila. BMC Evol Biol 15:175. https://doi.org/10.1186/s12862-015-0452-8
Kacsoh BZ, Bozler J, Ramaswami M, Bosco G (2015) Social communication of predator-induced changes in Drosophila behavior and germ line physiology. Elife 4:1–36. https://doi.org/10.7554/elife.07423
Kang L, Aggarwal DD, Rashkovetsky E et al (2016) Rapid genomic changes in Drosophila melanogaster adapting to desiccation stress in an experimental evolution system. BMC Genom 17:233. https://doi.org/10.1186/s12864-016-2556-y
Kerstes NA, Bérénos C, Schmid-Hempel P, Wegner KM (2012) Antagonistic experimental coevolution with a parasite increases host recombination frequency. BMC Evol Biol 12:18. https://doi.org/10.1186/1471-2148-12-18
Kilias G, Alahiotis SN, Onoufriou A (1979) The alcohol dehydrogenase locus affects meiotic crossing-over in Drosophila melanogaster. Genetica 50:173–177
Kimura M (1956) A model of a genetic system which leads to closer linkage by natural selection. Evolution (N Y) 10:278–287. https://doi.org/10.1111/j.1558-5646.1956.tb02852.x
King RC (1970) The meiotic behavior of the Drosophila oocyte. Int Rev Cytol 28:125–168. https://doi.org/10.1016/s0074-7696(08)62542-5
King RC, Aggarwal SK, Aggarwal U (1968) The development of the female Drosophila reproductive system. J Morphol 124:143–165. https://doi.org/10.1002/jmor.1051240203
Kohl KP, Singh ND (2018) Experimental evolution across different thermal regimes yields genetic divergence in recombination fraction but no divergence in temperature associated plastic recombination. Evolution (N Y) 72:989–999. https://doi.org/10.1111/evo.13454
König A, Shcherbata HR (2015) Soma influences GSC progeny differentiation via the cell adhesion-mediated steroid-let-7-Wingless signaling cascade that regulates chromatin dynamics. Biol Open 4:285–300. https://doi.org/10.1242/bio.201410553
Korol AB, Preygel IA, Preygel SI (1994) Recombination variability and evolution. Chapman & Hall, London
Laws KM, Drummond-Barbosa D (2017) Control of germline stem cell lineages by diet and physiology. In: Arur S (ed) Signaling-mediated control of cell division. Springer, Cham, pp 67–99
Mandal RK, Kwon YM (2017) Global screening of Salmonella enterica serovar Typhimurium genes for desiccation survival. Front Microbiol 8:1723
Markow TA, Matzkin LM, Watts TD (2007) Desiccation resistance in four Drosophila species: sex and population effects. Fly (Austin) 1:268–273. https://doi.org/10.4161/fly.5293
McClintock B (1984) The significance of response of the genome to challenge. Science 226:792–801. https://doi.org/10.1126/science.15739260
Modliszewski JL, Copenhaver GP (2017) Meiotic recombination gets stressed out: CO frequency is plastic under pressure. Curr Opin Plant Biol 36:95–102. https://doi.org/10.1016/j.pbi.2016.11.019
Neel JV (1941) A relation between larval nutrition and the frequency of crossing over in the third chromosome of Drosophila melanogaster. Genetics 26:506–516
Nei M (1967) Modification of linkage intensity by natural selection. Genetics 57:625–641
Parisi MJ, Gupta V, Sturgill D et al (2010) Germline-dependent gene expression in distant non-gonadal somatic tissues of Drosophila. BMC Genom 11:346. https://doi.org/10.1186/1471-2164-11-346
Plough HH (1917) The effect of temperature on crossingover in Drosophila. J Exp Zool 24:147–209. https://doi.org/10.1104/pp.4.2.281
Plough HH (1921) Further studies on the effect of temperature on crossing over. J Exp Zool 32:187–202. https://doi.org/10.1002/jez.1400320202
Politzer O (1940) Veränderungen der Crossoverhäufigkeit Durch Einwirkung von Temperatur und Alter (Versuche am III. Chromosom der Drosophila melanogaster). Z Indukt Abstamm Vererbungsl 78:129–147
Rajpurohit S, Gefen E, Bergland AO et al (2018) Spatiotemporal dynamics and genome-wide association analysis of desiccation tolerance in Drosophila melanogaster. Mol Ecol 27:3525–3540. https://doi.org/10.1111/mec.14814
Rybnikov SR, Frenkel ZM, Korol AB (2017) What drives the evolution of condition-dependent recombination in diploids? Some insights from simulation modelling. Philos Trans R Soc B 372:20160460. https://doi.org/10.1098/rstb.2016.0460
Rybnikov SR, Frenkel ZM, Fahima T, Korol AB (2018a) The evolutionary advantage of condition-dependent recombination in a Red Queen model with diploid antagonists. bioRxiv 478966:1–16. https://doi.org/10.1101/478966
Rybnikov SR, Frenkel ZM, Korol AB (2018b) Fitness-dependent recombination can be evolutionarily advantageous in diploids under mutation-selection balance. bioRxiv 381228:1–21. https://doi.org/10.1101/381228
Rzezniczak TZ, Merritt TJS (2012) Interactions of NADP-reducing enzymes across varying environmental conditions a model of biological complexity. G3: Genes Genomes Genet 2:1613–1623. https://doi.org/10.1534/g3.112.003715
Séguéla-Arnaud M, Crismani W, Larchevêque C et al (2015) Multiple mechanisms limit meiotic crossovers: TOP3α and two BLM homologs antagonize crossovers in parallel to FANCM. Proc Natl Acad Sci USA 112:4713–4718. https://doi.org/10.1073/pnas.1423107112
Segura J, Ferretti L, Ramos-Onsins S, Capilla L, Farre M, Reis F, Oliver-Bonet M, Fernandez-Bellon H, Garcia F, Garcia-Caldes M, Robinson TJ, Ruiz- Herrera A (2013) Evolution of recombination in eutherian mammals: insights into mechanisms that affect recombination rates and crossover interference. Proc Roy Soci B Biol Sci 280(1771):20131945–20131945
Shaw FH, Baer CF (2011) Fitness-dependent mutation rates in finite populations. J Evol Biol 24:1677–1684. https://doi.org/10.1111/j.1420-9101.2011.02320.x
Singh ND (2019) Wolbachia infection associated with increased recombination in Drosophila. G3: Genes Genomes Genet 9:229–237. https://doi.org/10.1534/g3.118.200827
Singh ND, Criscoe DR, Skolfield S et al (2015) Fruit flies diversify their offspring in response to parasite infection. Science 349:747–750
Stapley J, Feulner PGD, Johnston SE et al (2017) Variation in recombination frequency and distribution across eukaryotes: patterns and processes. Philos Trans R Soc B 372:20160455. https://doi.org/10.1098/rstb.2016.0455
Stern C (1926) An effect of temperature and age on crossing-over in the first chromosome of Drosophila melanogaster. Proc Natl Acad Sci USA 12:530–532
Stevison LS, Sefick S, Rushton C, Graze RM (2017) Recombination rate plasticity: revealing mechanisms by design. Philos Trans R Soc London B 372:20160459. https://doi.org/10.1098/rstb.2016.0459
Szauter P (1984) An analysis of regional constraints on exchange in Drosophila melanogaster using recombination-defective meiotic mutants. Genetics 106:45–71
Telonis-Scott M, Sgro CM, Hoffmann AA, Griffin PC (2016) Cross-study comparison reveals common genomic, network, and functional signatures of desiccation resistance in Drosophila melanogaster. Mol Biol Evol 33:1053–1067. https://doi.org/10.1093/molbev/msv349
Thaler DS (1994) The evolution of genetic intelligence. Science 264:224–225
Thorat L, Oulkar D, Banerjee K et al (2017) High-throughput mass spectrometry analysis revealed a role for glucosamine in potentiating recovery following desiccation stress in Chironomus. Sci Rep 7:3659. https://doi.org/10.1038/s41598-017-03572-5
Verde LA (2003) The effect of stress on meiotic recombination in maize (Zea mays L.). Iowa State University
Wexler Y, Rokhlenko O (2007) Prisoner’s dilemma posed by fitness-associated recombination strategies. J Theor Biol 247:1–10. https://doi.org/10.1016/j.jtbi.2007.01.031
Woodruff RC, Thompson JN (1977) An analysis of spontaneous recombination in Drosophila melanogaster males. Heredity (Edinb) 38:291–307. https://doi.org/10.1038/hdy.1977.92
Zetka MC, Rose AM (1995) Mutant rec-1 eliminates the meiotic pattern of crossing over in Caenorhabditis elegans. Genetics 141:1339–1349
Zhang L, Liang Z, Hutchinson J, Kleckner N (2014) Crossover patterning by the beam-film model: analysis and implications. PLoS Genet 10:e1004042. https://doi.org/10.1371/journal.pgen.1004042
Zhong W, Priest NK (2011) Stress-induced recombination and the mechanism of evolvability. Behav Ecol Sociobiol 65:493–502. https://doi.org/10.1007/s00265-010-1117-7
Zhuchenko AA, Korol AB (1985) Recombination in evolution and breeding. Nauka, Moscow
Zhuchenko AA, Korol AB, Gavrilenko TA, Kibenko TY (1986) The relation between genotype adaptivity and reactivity of its recombination characteristics under temperature effect. Genetika 22:966–974 (in Russian)
Zickler D, Kleckner N (2016) A few of our favorite things: pairing, the bouquet, crossover interference and evolution of meiosis. Semin Cell Dev Biol 54:135–148. https://doi.org/10.1016/j.semcdb.2016.02.024
Acknowledgments
We are grateful to two anonymous reviewers for critical comments and corrections which allowed to improve the manuscript. We also thank Kostas Illiadi, Eugene Kandel and Marios Kyriazis for fruitful discussions on soma-germline interactions.
Funding
The study was supported by the Israel Science Foundation (grant 1844/17); the University Grants Commission, India (DS Kothari project grant F.4-2/2006(BSR)/BL/16-17/0330); the Council for Higher Education of the Israeli Ministry of Education; and the Israeli Ministry of Aliyah and Integration.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Aggarwal, D.D., Rybnikov, S., Cohen, I. et al. Desiccation-induced changes in recombination rate and crossover interference in Drosophila melanogaster: evidence for fitness-dependent plasticity. Genetica 147, 291–302 (2019). https://doi.org/10.1007/s10709-019-00070-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10709-019-00070-6