Abstract
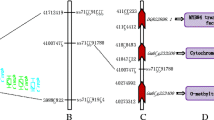
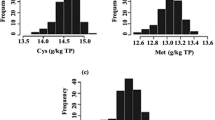
Soybean is important throughout the world due to its high seed protein and oil, while the quality and quantity of seed amino acids need to be improved. To improve the multiple amino acid concentrations in soybean simultaneously, detecting and utilizing the pleiotropic quantitative trait loci (QTL) and related genes become increasingly important. In view of this, a F6:7 recombinant inbred line population was genotyped using 1739 polymorphic SNP and 127 SSR markers in the present study and was phenotyped for seventeen types of amino acids simultaneously. In total, twelve co-located or overlapped pleiotropic additive QTL clusters, which explained 2.38–16.79% of the amino acid variation, were identified. Of them, one novel pleiotropic QTL cluster with a phenotypic variation explained ranging from 8.84 to 16.79% for ten kinds of amino acid contents (glycine, alanine, isoleucine, leucine, valine, methionine, aspartic acid, glutamic acid, lysine and phenylalanine), was located at the same position on linkage group D2, and the confidence interval was only 0.8 cM. Moreover, the individuals in this family-based population (345 lines) and another cultivar-based population (254 varieties) with different genotypes at the common flanking markers for this QTL cluster showed significantly different amino acid contents, which further validated the QTL mapping results. Additionally, some candidate genes that might participate in the amino acid biosynthesis process were found in these pleiotropic QTL regions. Thus, novel pleiotropic QTL clusters could be applied in marker-assisted selection breeding or map-based candidate gene cloning in soybean for multiple amino acid genetic improvements in seed in the future.
Similar content being viewed by others
References
Akond M, Liu S, Kantartzi SK, Meksem K, Bellaloui N, Lightfoot DA, Yuan J, Wang D, Anderson J, Kassem MA (2015) A SNP genetic linkage map based on the ‘Hamilton’ by ‘Spencer’ recombinant inbred line population identified QTL for seed isoflavone contents in soybean. Plant Breed 134(5):580–588
Azevedo RA, Lancien M, Lea PJ (2006) The aspartic acid metabolic pathway, an exciting and essential pathway in plants. Amino Acids 30(2):143–162
Carlson CM (2011) Genetic control of protein and amino acid content in soybean determined in two genetically connected populations. Dissertation, North Carolina State University
Chen Q, Yan L, Lei YK, Zhang YJ, Di R, Liu BQ, Yang CY, Zhang MC (2015) QTL mapping of seed amino acids content in soybean. Soybean Sci 34(4):597–600
Csanadi G, Vollmann JG, Lelley T (2001) Seed quality QTLs identified in a molecular map of early maturing soybean. Theor Appl Genet 103(6–7):912–919
Diers BW, Keim P, Fehr WR, Shoemaker RC (1992) RFLP analysis of soybean seed protein and oil content. Theor Appl Genet 83(5):608–612
Doyle J (1990) Isolation of plant DNA from fresh tissue. Focus 12:13–15
Fallen BD, Hatcher CN, Allen FL, Kopsell DA, Saxton AM, Chen PY, Kantartzi SK, Cregan PB, Hyten DL, Pantalone VR (2013) Soybean seed amino acid content QTL detected using the universal soy linkage panel 1.0 with 1,536 SNPs. J Plant Genome Sci 1(3):68–79
Ferreira RR, Varisi VA, Meinhardt LW, Lea PJ, Azevedo RA (2005) Are high-lysine cereal crops still a challenge? Braz J Med Biol Res 38(7):985–994
Friedman M, Brandon DL (2001) Nutritional and health benefits of soy proteins. J Agric Food Chem 49(3):1069–1086
He XL, Zhang JZ (2006) Toward a molecular understanding of pleiotropy. Genetics 173(4):1885–1891
Hwang EY, Song QJ, Jia GF, Specht JE, Hyten DL, Costa J, Cregan PB (2014) A genome-wide association study of seed protein and oil content in soybean. BMC Genom 15(1):1
Inc Soytech (2004) Oilseed statistics. Soytech Inc., Bar Harbor
Khandaker L, Akond M, Liu SM, Kantartzi SK, Meksem K, Bellaloui N, Lightfoot DA, Kassem MA (2015) Mapping of QTL associated with seed amino acids content in “MD96-5722” by “Spencer” RIL population of soybean using SNP markers. Food Nutr Sci 6(11):974–984
Li HH, Ribaut JM, Li ZL, Wang JK (2008) Inclusive composite interval mapping (ICIM) for digenic epistasis of quantitative traits in biparental populations. Theor Appl Genet 116(2):243–260
Li B, Tian L, Zhang JY, Huang L, Han FX, Yan SR, Wang LZ, Zheng HK, Sun JM (2014) Construction of a high-density genetic map based on large-scale markers developed by specific length amplified fragment sequencing (SLAF-seq) and its application to QTL analysis for isoflavone content in Glycine max. BMC Genom 15(1):1086
Li XH, Kamala S, Tian R, Du H, Li WL, Kong YB, Zhang CY (2018) Identification and validation of quantitative trait loci controlling seed isoflavone content across multiple environments and backgrounds in soybean. Mol Breed 38:8
Liu HY, Quampah A, Chen JH, Li JR, Huang ZR, He QL, Zhu SJ, Shi CH (2013) QTL mapping based on different genetic systems for essential amino acid contents in cottonseeds in different environments. PLoS ONE 8(3):e57531
Mukhtarov AS (2000) Analysis of a quantitative trait locus allele from wild soybean that increases seed protein concentration in soybean. Crop Sci 40(5):1438–1444
Nyquist WE, Baker RJ (1991) Estimation of heritability and prediction of selection response in plant populations. Crit Rev Plant Sci 10(3):235–322
Panthee DR, Pantalone VR, Sams CE, Saxton AM, West DR, Orf JH, Killam AS (2006a) Quantitative trait loci controlling sulfur containing amino acids, methionine and cysteine, in soybean seeds. Theor Appl Genet 112(3):546–553
Panthee DR, Pantalone VR, Saxton AM, West DR, Sams CE (2006b) Genomic regions associated with amino acid composition in soybean. Mol Breed 17(1):79–89
Qiu HM, Li Z, Yu Y, Gao SQ, Ma XP, Zheng YH, Meng FF, Hou YL, Wang YQ, Wang SM (2015) Mining and analysis of genes related to sulfur-containing amino acids in soybean based on Meta-QTL. Chin J Oil Crop Sci 37(2):141–147
Shen B, Li CJ, Tarczynski MC (2002) High free-methionine and decreased lignin content result from a mutation in the Arabidopsis s-adenosyl-l-methionine synthetase 3 gene. Plant J 29(3):371–380
Siehl DL (1999) The biosynthesis of tryptophan, tyrosine, and phenylalanine from chorismate. In: Singh BK (ed) Plant amino acids: biochemistry and biotechnology. Marcel Dekker Inc., New York, pp 171–204
Singh BK (1999) Biosynthesis of valine, leucine, and isoleucine. In: Singh BK (ed) Plant amino acids: biochemistry and biotechnology. Marcel Dekker Inc., New York, pp 227–247
Tian R (2017) QTL mapping of amino acid content in soybean seed. Dissertation, Hebei Agricultural University
Vaughn JN, Nelson RL, Song QJ, Cregan PB, Li ZL (2014) The genetic architecture of seed composition in soybean is refined by genome-wide association scans across multiple populations. Genes, Genom Genet 4(11):2283–2294
Warrington CV (2011) QTL mapping and optimum resource allocation for enhancing amino acid content in soybean. Dissertation, University of Georgia
Warrington CV, Abdel-Haleem H, Hyten DL, Cregan PB, Orf JH, Killam AS, Bajjalieh N, Li Z, Boerma HR (2015) QTL for seed protein and amino acids in the Benning × Danbaekkong soybean population. Theor Appl Genet 128(5):839–850
Funding
This research was funded by the Project of Hebei province Science and Technology Support Program (16227516D-1, 17927670H) and the Fund of Research Group Construction for Crop Science in Hebei Agricultural University (TD2016C203).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors have declared that no competing or conflicts of interest exist.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Li, X., Tian, R., Kamala, S. et al. Identification and verification of pleiotropic QTL controlling multiple amino acid contents in soybean seed. Euphytica 214, 93 (2018). https://doi.org/10.1007/s10681-018-2170-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10681-018-2170-y