Abstract
A variety of pseudomonads are associated with diseases of Actinidia and Prunus plants. A recently emerged virulent haplotype of Pseudomonas syringae pv. actinidiae (biovar 3) causes severe stem cankers on kiwifruit associated with dieback of canes. Pseudomonas syringae also causes one of the most important bacterial diseases affecting cherry orchards worldwide. Bacterial canker of cherry limits production in orchards in New Zealand. Less virulent and non-pathogenic pseudomonads also exist on both hosts, providing an opportunity to investigate the diversity of pseudomonads on these hosts, to find identifiers for new emerging highly pathogenic strains that could be used at the border to prevent incursions. In this study, genetic typing was used to explore the diversity of Pseudomonas on kiwifruit and a variety of Prunus plants. Multilocus sequence analysis (MLSA) of four house-keeping genes separated pseudomonads from kiwifruit and stone fruit plants into two major groups: one corresponding to P. syringae sensu lato and one corresponding to other Pseudomonas species. Within P. syringae sensu lato, strains were assigned to six of the nine previously described genomospecies or five of the seven previously described phylogroups. In the other major group, strains from both hosts clustered with a variety of well characterised non-pathogenic pseudomonads. The classification of strains into the two major groups is of practical diagnostic value since the most common pathogens of fruit trees belong to the P. syringae species complex. Furthermore, our analysis suggests that molecular diagnostics might be possible for the classification of strains into these two groups as a first tool to screen for exotic pathogenic pseudomonads on germplasm imports of both commodities.
Similar content being viewed by others
Introduction
Pseudomonads cause disease on a wide range of monocotyledonous and dicotyledonous plants worldwide. In the Pseudomonas syringae species complex alone, also referred to as P. syringae sensu lato, there are over 60 pathovars that cause a range of blight, speck, spot, and canker diseases (Dye et al. 1980; Agrios 1997; Young 2010). Pathovars of P. syringae responsible for economic damage to commercially important crop plants have been the major focus of studies associated with this species complex (Hirano and Upper 1990; Morris et al. 2009; Lamichhane et al. 2014). However, strains of P. syringae have not only been isolated from cultivated plants but also from air, clouds, soil, water and snow (Goode and Sasser 1980; Shaffer and Lighthart 1997; Amato et al. 2006; Morris et al. 2008), leaf litter (Monteil et al. 2012; Tyson et al. 2012), and non-host plants adjacent to crops (Lindemann et al. 1984; Jakob et al. 2002; Vicente et al. 2004).
A range of pathogenic pseudomonads have previously been isolated from the botanically diverse hosts, kiwifruit (Actinidia spp.) and stone fruits (Prunus spp.). Most notable of these, in terms of disease epidemics, were Pseudomonas sp. (blossom blight) and P. s. pv. actinidiae from kiwifruit, P. s. pv. morsprunorum from stone fruit and P. syringae pv. syringae from both hosts. Consequently, much of the research has focused on these important pathogens (Crosse 1966; Young et al. 1997; Balestra and Varvaro 1997; Hu et al. 1999; Balestra et al. 2008a; Bultreys and Kaluzna 2010).
On kiwifruit, P. s. pv. syringae and Pseudomonas sp. (Young et al. 1997; Hu et al. 1999) infect buds and flowers causing moderate to severe blossom loss under favourable environmental conditions (Everett and Henshal 1994; Balestra and Varvaro 1997; Balestra et al. 2008a). However, although P. s. pv. syringae could be isolated from diseased blossoms in New Zealand orchards, the populations were far fewer than those of Pseudomonas sp., and thus were probably present as a secondary invader (Everett and Henshal 1994). P. s. pv. syringae and Pseudomonas sp. have been ascribed as the cause of blossom blight disease symptoms in Portuguese, Chinese, Japanese and Korean orchards (Ieki 1993; Miyoshi and Tachibana 1996; Hu et al. 1998; Koh et al. 2001; Balestra et al. 2009), and P. fluorescens in Korea (Lee et al. 2009). Other studies have shown that Pseudomonas sp. and P. s. pv. syringae were found in blossoms in Italian orchards infected with P. s. pv. actinidiae. These authors hypothesied that all three Pseudomonads were present as epiphytes (Purahong et al. 2018) and eventually form a syndemic association to cause disease.
Pseudomonas syringae pv. actinidiae (Psa) is the causal agent of bacterial canker of kiwifruit (Takikawa et al. 1989). A recent and emergent virulent haplotype of P. s. pv. actinidiae (biovar 3) (Chapman et al. 2012; McCann et al. 2013; Vanneste et al. 2013) causes leaf spots, rotting of flowers, bud rot, severe stem cankers, cane dieback and oozing of red or white exudates (Balestra et al. 2008b; Everett et al. 2011; Mazzaglia et al. 2012). A total of four biovars of P. syringae pv. actinidiae affecting kiwifruit were described until 2014, which included biovar 4 that later was redefined as P. syringae pv. actinidifoliorum by Cunty et al. (2015). More recently, two new biovars, biovars 5 and 6, have been reported from Japan as being closely related to the Korean Psa biovar 2 (biovar 5), and the global pandemic biovar 3 (biovar 6) (Scortichini et al. 2012; Fujikawa and Sawada 2016; Morán et al. 2018; Fujikawa and Sawada 2019).
Among the main bacterial pathogens affecting stone fruits worldwide are P. s. pv. morsprunorum (race 1 and 2) and P. s. pv. syringae, both inducing bacterial canker in sweet and sour cherry (Prunus avium and P. cerasus) (Agrios 1997; Bultreys and Kaluzna 2010). A closely related pathogen, P. s. pv. persicae, causes a decline symptom, resulting in tip dieback and necrosis of branches in peach, nectarine and Japanese plum in New Zealand (Young 1987). More recently, strains isolated from diseased sour cherry tissue in Poland were described as Pseudomonas cerasi species novel causing symptoms resembling those of bacterial canker (Kałużna et al. 2016). In the UK, the incidence of bacterial canker by P. s. pv. syringae and P. s. pv. morsprunorum was reported to be 50–100% on wild cherry (Vicente et al. 2004). In Europe, other highly aggressive P. syringae pathovars are known to be responsible for causing serious diseases on other woody plants such as horse chestnut (Aesculus hippocastanum) and hazelnut (Corylus avellana L.), caused by P. s. pv. aesculi and P. s. pv. coryli respectively, which pose major threats to agroforestry (Lamichhane et al. 2014). Accordingly, the focal point of research has been on these economically important bacterial pathogens.
Other, apparently less aggressive or non-virulent, pseudomonads also exist in kiwifruit orchards, with epiphytic P. syringae and P. fluorescens strains isolated or detected from leaf and blossom surfaces (Everett and Henshall1994; Purahong et al. 2018; Straub et al. 2018), but little is known about them. However, pioneering work on cherry trees in England had shown the possibility of epiphytic growth of the pathogen P. s. pv. morsprunorum on healthy leaves (Crosse 1963; Crosse 1966). This was later confirmed by the study of epidemiology of P. syringae on various hosts including deciduous fruit trees, tomato, ornamental trees and beans (Hirano and Upper 1990). Not only does P. syringae grow epiphytically on leaf surfaces, but it also invades through the stomata to grow in the apoplast without causing disease symptoms.
Phenotypic LOPAT (Levan production, Oxidase reaction, Pectolytic activity on potato, Arginine dehydrolase activity, and the ability to cause a hypersensitive response (HR) on tobacco) tests have traditionally been used to type P. syringae isolates (Lelliott et al. 1966). More recently, multilocus sequence analysis (MLSA) (Maiden et al. 1998) has proven to be a powerful approach for the differentiation of strains and clonal lineages of pseudomonads as well as being used to ascertain their relatedness (Sarkar and Guttman 2004; Bull et al. 2011; Chapman et al. 2012; Berge et al. 2014; Glaeser and Kämpfer 2015; Gomila et al. 2015; Kałużna et al. 2016; Straub et al. 2018).
Recently other workers have examined Psa isolates from kiwifruit and compared these with environmental pseudomonads and pseudomonads from woody hosts including Prunus domestica (plum) and P. armenica (apricot) using MLSA (Bartoli et al. 2015). This work led to the identification of gene regions that were used to design polymerase chain reaction (PCR) primers that could distinguish between phylogroups of Pseudomonas syringae (Borschinger et al. 2016). Furthermore, this analysis showed that strains isolated from the environment could be pathogenic on economically important hosts (Bartoli et al. 2015). Our current study examines the populations of pseudomonads isolated from kiwifruit, cherry and apricot. In contrast to those earlier studies, our study aimed to distinguish saprotrophic and pathogenic strains from the same host.
To this end, LOPAT and MLSA were used to define the diversity of a collection of pseudomonads isolated from kiwifruit and a variety of Prunus species (P. avium, P. armeniaca, P. cerasus, P. dulcis P. persica and P. x yedoensis) in New Zealand and overseas. These data provide novel insights into the diversity of pseudomonads associated with kiwifruit and Prunus plants, two economically important commodities for New Zealand trade. The diversity of pseudomonad strains on both hosts poses significant practical issues for defining the threat of exotic strains to the kiwifruit and various stone fruit industries as part of border biosecurity and pest management. This study was initiated to find genetic differences that could be used to distinguish strains that are pathogenic, and likely to pose a biosecurity threat to economically important plants, from non-pathogenic pseudomonads.
Materials and methods
Bacterial strains and culture conditions
All bacterial strains (Table 1) were routinely grown on Pseudomonas F agar (Difco™) for 48 h or in Luria Bertani (LB) broth for 16 h, at 28 °C. For long-term storage, strains were kept as LB-glycerol stocks at -80 °C. The majority of strains were obtained as cultures from the International Collection of Microbes from Plants (ICMP) Landcare Research, Auckland, New Zealand (http://www.landcareresearch.co.nz/resources/collections/icmp). The remainder were acquired as cultures from the New Zealand Institute for Plant and Food Research Limited (PFR) including cultures from the Ministry for Primary Industries (MPI) collections. These isolates were obtained from different organs (leaves, buds, flowers, etc.) of Actinidia and Prunus plants in New Zealand and other countries, as indicated in Table 1. Many of the strains in the collections chosen for this study were not fully characterised to the pathovar level at the time of collection, their taxonomic classification remaining to genus or species level. A number of well-characterised pathogenic and non-pathogenic Pseudomonas species were also included in this study for comparative purposes.
LOPAT tests
LOPAT tests were performed as originally described (Lelliott et al. 1966) with minor modifications to the tests for oxidase activity. The agar used for the levan test was nutrient agar (Difco) containing 5% sucrose. For the oxidase test, paper strips containing Kovacs’ reagent (Oxoid™) were used instead of traditional filter paper flooded with N,N-dimethyl-p-phenylenediamine. Strips for the oxidase reaction were smeared with a full loop of fresh culture, following the manufacturer’s instructions. LOPAT tests were carried out in at least three independent experiments.
MLSA
Genomic DNA from bacterial isolates was purified using the Qiagen™ Blood-tissue DNA extraction kit, following the manufacturer’s instructions for Gram-negative bacteria. Partial sequences for gapA, gltA, gyrB and rpoD were amplified from the DNA of each isolate by PCR, using the primer pairs described by Hwang et al. (2005). Each PCR was carried out in a total volume of 20 μL containing final concentrations of 1× PCR Buffer (20 mM Tris-HCl [pH 8.4] and 50 mM KCl), 0.2 mM of each dNTP, 0.25 μM of each primer, 2 mM MgCl2 and 0.05 U of Invitrogen Taq polymerase (Invitrogen, Carlsbad, CA). Approximately 50 to100 ng of template DNA was also used in each reaction. PCR conditions included an initial denaturation step at 94 °C for 2 min followed by 30 cycles of amplification, each cycle consisting of template denaturation at 94 °C for 2 min, annealing at 56 °C for 1 min and extension at 72 °C for 1 min, and a final extension step at 72 °C for 7 min. Purification of each PCR product was performed using the Qiagen™ PCR purification kit and DNA sequencing of the product was carried out in both directions by Macrogen Inc. DNA sequences were trimmed to the same lengths as used by Hwang et al. (2005). These were 476 bp for gapA, 529 bp for gltA, 495 bp for rpoD and 507 bp for gyrB. The concatenated sequence for each isolate was then submitted to the Plant Associated and Environmental Microbes Database (PAMDB) (www.pamdb.org; (Almeida et al. 2010)).
Concatenated DNA sequences for gapA, gltA, gyrB and rpoD from each isolate were compared with those for type strains and other well-characterised isolates of Pseudomonas, downloaded from PAMDB (Table S1). These included representative strains from eight of the nine previously defined genomospecies of Pseudomonas (Gardan et al. 1999), and all seven of the phylogroups designated by (Parkinson et al. 2011). Strains from other hosts rather than kiwifruit and Prunus spp. is presented in Table S1.The concatenated sequences of the four loci were aligned using MUSCLE (Edgar 2004), and a maximum likelihood tree was constructed using the general time reversible (GTR) substitution model as per Bull et al. (2011) in Geneious version 10.0.9.1 (Guindon et al. 2010; Drummond et al. 2013). A total of 1000 bootstrap replicates were performed to measure confidence in the branch points.
To validate the identification of strains of P. viridiflava in our collection by MLSA, a second analysis was performed using the concatenated sequences for just the gapA and gyrB genes. The comparison of these concatenated DNA sequences with those from a variety of putative P. viridiflava isolates analysed by Bartoli et al. (2014), was performed as described above for MLSA using gapA, gltA, gyrB and rpoD.
Results
DNA analysis
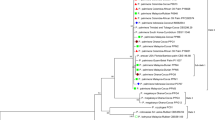
MLSA divided the concatenated DNA sequences, which included four gene regions (gyrB, gapA, gltA and rpoD) of a total of 101 pseudomonad strains (33 from Actinidia spp., 24 from Prunus and 44 from other hosts downloaded from GenBank), into two distinct groups (Group A and Group B) (Fig. 1). Group A corresponded to P. syringae sensu lato and included 22 strains from kiwifruit (Actinidia chinensis var. chinensis and A. chinensis var. deliciosa) and 19 strains from Prunus spp. (P. avium, P. armeniaca, P. cerasus, P. dulcis, P. persica and P. x yedoensis), while Group B consisted of other Pseudomonas species. Group A isolates were diverse, clustering into six of the nine genomospecies of P. syringae described by Gardan et al. (1999), and into five of the seven phylogroups previously recognised by Parkinson et al. (2011) as constituting the P. syringae species complex, or the thirteen phylogroups later defined by Berge et al. (2014) (Table 1; Fig. 1). Group A also included a variety of well-characterised phytopathogenic isolates of P. syringae such as P. syringae pv. tomato (strain Pto DC3000) or P. syringae pv. phaseolicola (strain Pph 1448A) as well as the type strains for P. viridiflava (ICMP 2848) and P. cannabina (CFBP 2341).
Multilocus sequence analysis (MLSA) of pseudomonads isolated from kiwifruit and stone fruits. An unrooted Maximum Likelihood tree (Guindon et al. 2010) constructed using concatenated DNA sequences of gapA, gltA, gyrB and rpoD from Pseudomonas collected from kiwifruit (green), stone fruits (red) and other hosts (black). Type strains for different pathovars are designated by T. Pathotype strains are designated by PT. Phylogroups (P) are designated where appropriate. On the right, grey bars were used to show two groups of P. syringae strains that cause canker and bud-rot in kiwifruit, and a third bar for P. s. pv. syringae that causes canker in sweet cherry. Bootstrap values >70% from 1000 replicates are indicated above the nodes
Group A
Phylogroup I
MLSA grouped five strains from Prunus spp. and six strains from kiwifruit together within phylogroup I. Three types of P. s. pv. actinidiae were in this phylogroup, those originally described as Psa-V or Psa-3 (virulent) (Chapman et al. 2012; McCann et al. 2013), which have since also been referred to as biovar 3 (Vanneste et al. 2013); those originally described as Psa-LV (low virulence), Psa 4 or biovar 4 (Chapman et al. 2012; McCann et al. 2013; Vanneste et al. 2013), which have now been assigned to a new pathovar, P. s. pv. actinidifoliorum (Cunty et al. 2015) and strain NCPPB 3739 PT, biovar 2 (Ferrante and Scortichini 2009). Strains ICMP 11292 isolated from kiwifruit bud and ICMP 8406 from peach together with a strain from apricot, P. s. pv. persicae CFBP 1573, segregated in a sub-clade together with strain Pto DC3000 from tomato and pathotype strains from other hosts: P. syringae pv. apii - Pap CFBP 2103 PT (celery) and P. syringae pv. maculicula - Pma CFBP 1657 PT (cabbage). Strains from Prunus - P. syringae ICMP 3690, P. s. pv. morsprunorum ATCC 19322 and the pathotype strain P. s. pv. avellanae NCPPB 3487 - also clustered in phylogroup I.
Phylogroup II
Pseudomonas syringae pv. syringae strains isolated from sweet cherry cankers aligned in phylogroup II, in a well-supported sub-clade together with P. syringae strains ICMP 13102, PD 2774 and ICMP 11168 isolates from kiwifruit and P. syringae ICMP 4122 from apricot. Isolates from kiwifruit, P. syringae strains ICMP 10191, ICMP 11293 and NCPPB 3871 segregated together in a sub-clade in phylogroup II. A recently described novel species of P. cerasi in Poland (Kałużna et al. 2016), from sour cherry diseased tissue, also clustered within strains of phylogroup II.
Phylogroup III
Phylogroup III contained ICMP 13303 and ICMP 3272, deposited initially into the ICMP culture collection as P. viridiflava after isolation from necrotic flower buds of A. chinensis var. chinensis and A. chinensis var. deliciosa in New Zealand, together with other P. syringae strains, defining a subset that comprises mostly bud-rot producing Pseudomonas closely related to P. s. pv. savastanoi. It also contained an unusual isolate of P. syringae ABAC10 isolated from kiwifruit leaves, which was LOPAT 1a. All other P. syringae LOPAT 1a from kiwifruit (ICMP 11168, ICMP 13102, PD 2774) were in Phylogroup II.
An additional tree constructed with concatenated gapA and gyrB sequences (Fig. S1) and including more sequences downloaded from GenBank than were in the four-gene analysis, confirmed that ICMP 3272 and ICMP 13303, isolated from blossom blight symptoms on kiwifruit (Table 1), clustered with P. s. pv. savastanoi in phylogroup III.
The second subset in phylogroup III was formed by a variety of P. syringae that are defined as pseudomonads that infect woody plants (Lamichhane et al. 2014). In this subset, our isolates from Prunus spp., such as Pseudomonas amygdali ICMP 3918, P. pv. morsprunorum (ICMP 3676 and SBV_PS6), and two strains of P. s. pv. cerasicola (ICMP 17525 and ICMP 13929), grouped together with P. s. pv. aesculi NCPPB 3681 PT and P. s. pv. cerasicola CFBP 6109 PT. Other pseudomonads from different hosts such as P. s. pv. savastanoi LMG 2209 PT (olive), P. s. pv. phaseolicola 1448A (bean), P. s. pv. sesame LMG 2289 PT (sesame) and P. s. pv. tabacci NCPPB1427 PT (tobacco) belong to this phylogroup III previously defined by Parkinson et al. (2011).
Phylogroup V
Strain PD 2766 isolated from A. chinensis var. chinensis in the United States and ICMP 13104, an isolate originally described as P. viridiflava isolated from A. chinensis var. deliciosa in France, were placed within phylogroup V together with two strains of P. cannabina Pca CFBP 2341 T and Pca CFBP 1637, forming a sub-clade on their own. No isolate from stone fruit fell in this phylogroup.
Phylogroup VII
Phylogroup VII contained an atypical isolate from kiwifruit (ICMP 11289) that was originally called P. marginalis. This clade also contained P. viridiflava from peach (ICMP 8820), a pathotype strain of P. viridiflava from golden currant (LMG 2276) as well as the type strain of P. viridiflava from bean (ICMP 2848). Further analysis, including more strains from GenBank, confirmed that ABAC 43 and ICMP 11289 from kiwifruit and P. viridiflava ICMP 8820 from P. persica clustered together with the type strain ICMP 2848 from Phaseolus vulgaris, and with the remaining P. viridiflava strains from symptomless leaf material of various hosts in Phylogroup VII (Fig. S1, Table S2).
Group B
Nine pseudomonads from kiwifruit and five from Prunus clustered into Group B along with well-known non-pathogenic pseudomonads such as P. fluorescens SBW25 and P. putida GB1, and biocontrol agents, including P. brassicacearum Q8r1–96 (Stockwell et al. 2010; Loper et al. 2012). Importantly, Group B does not include any known pathogens of either kiwifruit or stone fruits.
ICMP 9505 was originally isolated post-harvest from kiwifruit and considered an opportunistic strain. ICMP 9505 was classified as P. marginalis based on previous LOPAT tests and clustered closely with the type strain, ICMP 3553, using MLSA in this study. Strain ICMP 3553 is the type strain for P. marginalis, which was originally isolated from chicory and subsequently shown to be pathogenic to chicory, chrysanthemum, tomato and celery (https://scd.landcareresearch.co.nz/Specimen/ICMP_3553). A strain of Pseudomonas sp. isolated from stem canker of P. persica, strain ICMP 460, clustered with ICMP 9505.
LOPAT tests
The analysed pseudomonads from kiwifruit and Prunus plants differed in several phenotypic traits according to their LOPAT profiles (Table 1). Twenty-seven were negative in the oxidase, potato soft rot and arginine dehydrolase assays, but produced an HR on tobacco indicative of LOPAT 1. Levan production was variable, grouping 22 strains into LOPAT 1A (levan positive) and five into LOPAT 1B (levan negative). Four strains presented the classical LOPAT 2 profile exemplified by P. viridiflava (absence of levan exopolysaccharide production, induction of HR on tobacco and soft rot of potato). Ten Pseudomonas strains were classified as LOPAT 2. ICMP 9505 was the only strain from kiwifruit with a LOPAT 4 profile. ICMP 9505, however, produced levan polysaccharide, while the type strain for P. marginalis (ICMP 3553, isolated from chicory) did not. This differentiated ICMP 9505 and ICMP 3553 into LOPAT 4A and 4B, respectively. A further six strains had a LOPAT 5 profile indicative of P. fluorescens (oxidase and arginine dihydrolase positive, but unable to induce soft rot on potato or a HR on tobacco). These strains could be divided into LOPAT 5A and 5B by virtue of variable production of levan. The four remaining strains produced non-typical LOPAT profiles that were not categorised by Lelliott et al. (1966). ABAC 63 was negative in all assays; ABAC 21 produced levan, but was negative in all other tests; ICMP 564 was oxidase positive only; and ICMP 460 was oxidase and potato soft rot positive, with the rest of tests being negative. An isolate from kiwiberry (Actinidia arguta), strain T14-1798A3, presented a LOPAT 3 profile.
Discussion
This study showed that irrespective of the host from which the Pseudomonas isolates had been recovered, the phylogenetic analysis of the concatenated DNA sequences of gapA, gltA, gyrB and rpoD placed isolates into two groups, designated A and B. Group A corresponded to P. syringae sensu lato and contained phylogenetic groups I, II, III, IV, V, VI and VII (Parkinson et al. 2011; Berge et al. 2014), and included pathogenic strains of pseudomonads. Group B contained mostly saprotrophs of other Pseudomonas species such as P. fluorescens, some strains with uncategorised LOPAT results, and one type strain of the pathogen P. marginalis pv. marginalis (ICMP 3553) isolated from chicory.
Although P. marginalis pv. marginalis can be pathogenic on some hosts, a review of the literature showed that the soft rot disease of chicory was caused by a disease complex consisting of two Erwinia spp. as well as P. marginalis ICMP 3553 (Schober and Zadoks 1999). From our results it is therefore possible that this bacterial isolate was associated with the symptoms, but not the cause, and was not a pathogen, and was thus correctly categorised in group B, non-pathogens.
Isolate ICMP 9505 was also classified as P. marginalis based on LOPAT tests, but was one of the nine Group B pseudomonads from kiwifruit. ICMP 9505 was originally isolated post-harvest from kiwifruit and considered an opportunistic strain. ICMP 9505 clustered closely with the type strain, ICMP 3553, using MLSA in this study. However, ICMP 3553 is also possibly an opportunistic strain, as discussed previously. Therefore it is possible that both strains are opportunistic weak pathogens or saprotrophs, explaining their presence in Group B. A strain of Pseudomonas sp. isolated from stem canker of P. persica, strain ICMP 460, also clustered with ICMP 9505 in Group B, and thus may also be an opportunistic invader.
LOPAT tests were used as determinative tests for identifying pseudomonads from plants since 1966 (Lelliott et al. 1966) and were all that were required for identification for several decades after (e.g. Everett and Henshall 1994; Marchi et al. 2005). However, our study has shown that LOPAT tests did not always agree with MLSA typing, suggesting that LOPAT tests are not as reliable as this more advanced technology. This agrees with recent studies comparing sequence-based typing with LOPAT tests, which showed that P. viridiflava strains have variable levan production and hypersensitivity response on tobacco (González et al. 2003; Bultreys and Kaluzna 2010; Sarris et al. 2012), and soft rotting ability (Bartoli et al. 2014).
In particular, ICMP 11289, initially isolated from buds and identified as P. marginalis, LOPAT 4B (Young and Fletcher 1997), was assigned to Pseudomonas sp. LOPAT 2 by our tests. The MLSA of this isolate did not place it with the other P. marginalis isolates ICMP 3553 or ICMP 9505 in Group B, nor with the other Pseudomonas sp. LOPAT 2 in phylogroup III. Instead, phenotyping this strain classified it as LOPAT 2, and based on MLSA it clustered with the type strain for P. viridiflava (ICMP 2848) in phylogroup VII of Group A.
Another exception, ICMP 561, isolated from P. avium, was initially deposited into the ICMP collection as P. s. pv. morsprunorum LOPAT 1A, but subsequently determined to belong to a Pseudomonas sp. because of an atypical LOPAT result in our study. This isolate clustered with Group B within the P. fluorescens/putida complex, thus confirming that it does not belong to P. s. pv. morsprunorum, and from this result is probably a saprotroph.
Our results therefore confirmed the conclusions by Berge et al. (2014) that the analysis of phenotypic traits provided by LOPAT groups did not correlate strongly with their classification into phylogroups by MLSA. For example, isolates designated as LOPAT 1 clustered into phylogroups I, II and III and isolates designated LOPAT 2 clustered into phylogroups III and VII. These results were consistent with those of Berge et al. (2014), who showed that few phylogroups possessed unique phenotypic patterns.
Thus the discrepancies between LOPAT results of the past and the present study, and the MLSA results, could be due to a number of factors. First, LOPAT results can be variable, even for the same species - as shown in Bartoli et al. (2014). The second possibility is that experimental error resulted in some cultures being mislabelled or contaminated. And the third possibility is that the new MLSA techniques are more reliable and accurate than LOPAT tests, as shown by Berge et al. (2014).
MLSA also has higher resolution than DNA-DNA hybridization or 16S rDNA sequencing for intraspecies variation (Sarris et al. 2012). Using only two of the gene regions (gyrB and gapA) confirmed the phylogroup assignation from analysis of four gene regions (gyrB, gapA, rpoD and gltA) of one of the difficult groups (P. viridiflava/Pseudomonas sp.), demonstrating the high resolution possible at the pathovar level, with only two gene regions. These pathogenic strains from kiwifruit were originally called P. viridiflava but have been simply designated Pseudomonas sp. because of their similarity to the savastanoi group, determined by plant and biochemical tests (Young et al. 1997; Hu et al. 1999). Based on MLSA, Berge et al. (2014) showed that phylogroup III contained isolates of P. s. pv. savastanoi, confirming the assignment of the kiwifruit pathogen to the savastanoi group. However, these isolates from kiwifruit were in a separate branch of this clade, recently defined by Straub et al. (2018) as a new subset (PG3a), and were clearly different from the other isolates from Prunus spp. and bean.
In contrast to the saprotrophs, the pathogenic strains from New Zealand (kiwifruit and Prunus) aligned well with the species phylogroups with which they had been previously identified (Sarkar and Guttman 2004; Rees-George et al. 2010; Parkinson et al. 2011; Berge et al. 2014). MLSA also grouped six isolates from kiwifruit and eight strains from stone fruits together within phylogroup III. This phylogroup comprises pathogens of host plants such as green beans (e.g. P. s. pv. phaseolicola and P. s. pv. glycinea) and is slightly separated from pathogens of woody plants (e.g. P. s. pv. aesculi, P. s. pv. savastanoi, P. amygdali). Two strains of P. s. pv. morsprunorum, one of the causal agents of canker in sweet cherry, belonged to this phylogroup and grouped with phylogroup III strains from woody plants. Although the kiwifruit isolates were separated in a third clade within phylogroup III, defining a subset of Pseudomonas isolates associated with bud rot, their close genetic relationship with vascular pathogens of woody plants suggests that they may have the capacity to cause vascular disease of kiwifruit. Frampton et al. (2014) suggested P. syringae ABAC10 and P. syringae pv. actinidiae were similar, based on the sensitivity of isolate ABAC10 to a large number of phages specific to P. syringae pv. actinidiae (Psa) – Biovar 3. Two new biovars of P. syringae pv. actinidiae - biovars 5 and 6 - were recently described in Japan by Fujikawa and Sawada (2016, 2019). These biovars 5 and 6 were found to be closely related to biovar 2 and biovar 3 respectively (Moran et al. 2018; Fujikawa and Sawada 2019), and clustered within phylogroup I with the remaining Psa strains.
The third clade within Phylogroup III contains P. syringae strains ICMP 19498 and ICMP 19499, described from a study of P. syringae by Straub et al. (2018) as phylogroup 3a (PG3a), containing commensal strains. Pseudomonas sp., also in PG3a, is pathogenic on kiwifruit although vascular symptoms have not been observed: only kiwifruit flower buds and leaves were affected (Wilkie et al. 1973). The severity of symptoms on buds was dependent on the stage of development when infection occurred; some were completely rotted and dropped, some partially opened then dropped, or some opened completely with only the stamens rotted, but developing fruit were deformed. Leaves were covered with angular necrotic spots (Young 1988). Although there was evidence that new infections were occurring during the season (Everett and Henshall1994), it is not known where the inoculum to start the epidemic in spring on this deciduous host overwintered. It is thus possible that Pseudomonas sp. may be systemically infecting kiwifruit vines, without observable symptoms. Recently, Straub et al. (2018) showed that a P. syringae strain of this subclade was able to colonize the leaf surface and apoplast of kiwifruit without producing visible disease symptoms, even though it was originally isolated from a leaf spot symptom. Pathogenic pseudomonads have an epiphytic stage to their life cycle, and the signal for parasitism is possibly the physical impact of rain (Hirano and Upper 1990), which was not simulated during laboratory inoculations. Other factors may also have contributed to the lack of symptoms following laboratory inoculation with this strain.
The results demonstrated that strains within the ‘P. cannabina cluster’ were not present within the isolates from kiwifruit and Prunus spp. considered in this study. This group included the type strain for this species (CFBP 2341) as well as the isolates PD 2766 and ICMP 13104 from the United States and France, defined as P. s. pv. syringae and P. viridiflava, respectively.
The non-pathogenic MLSA Group B isolates were genetically diverse, demonstrating that the Pseudomonas populations on kiwifruit and Prunus plants are complex. It has been proposed that the emergence of new pathogenic bacteria may be associated with the acquisition not only of virulence genes, but also of other traits (e.g. antimicrobial activity) that enable one strain to outcompete another (Garlant et al. 2013). If this is the case, the high diversity of non-pathogenic isolates capable of epiphytic and endophytic growth would add to the pool of genetic traits available to emerging pathogens through lateral gene transfer. Lateral gene transfer between related pseudomonads can occur readily as a result of selective pressure such as that imposed by plant host resistance, resulting in genome remodelling and changes in virulence (Pitman et al. 2005; Lovell et al. 2009; O'Brien et al. 2012).
Finally, it is important to note that the high diversity of pathogenic and non-pathogenic bacteria on plant hosts as described here poses significant practical issues for border biosecurity and pest management. However, recently developed PCR primers have been able to distinguish Pseudomonas syringae from all other species of pseudomonads (Guilbaud et al. 2016). Additionally, Bartoli et al. (2015) designed PCR primers based on the catechol operon genes that were found in pathogenic strains of Pseudomonas syringae but not in non-pathogenic strains. These primers failed to detect P. s. pv. syringae strains from cherry canker, but they did distinguish mildly pathogenic environmental strains from those that did not produce symptoms on kiwifruit, showing the potential of this approach. Environmental strains of P. syringae were assigned to phylogroup I by using the same four gene regions in MLSA as were used in the current study. However, environmental strains have been shown by Morris et al. (2008) to be part of the life cycle of pathogenic strains of pseudomonads, and are therefore not necessarily non-pathogens.
In contrast, our results showed that all pathogenic strains from both hosts were placed into Group A and the saprophytic strains into Group B, suggesting that sequence-based identification may have the potential for development into a PCR-based tool for phytosanitary procedures. These results suggest that markers could be designed to be used by biosecurity specialists using standard molecular tests, such as PCR or Loop-mediated isothermal amplification (LAMP) (Tomlinson and Boonham 2008; Harper et al. 2010), to resolve the pathogenic status of pseudomonad isolates from plant commodities quickly at the border. Our current challenge is to find some loci that can be unique to Group A or B, for which we started to analyse the genome sequences of selected strains (Visnovsky et al. 2016), and upon which a quick screening method using a simple molecular test could be based for post-entry quarantine (PEQ) testing.
In the meantime, the classification of strains into either a probably pathogenic or probably saprophytic group by MLSA could be applied as a first molecular screening tool for border security.
Change history
13 August 2019
This erratum has been created as many author corrections were overlooked in proofing stage.
References
Agrios, G. N. (1997). Plant diseases caused by prokaryotes: Bacteria and mollicutes. Plant Pathology, 4, 407–470.
Almeida, N. F., Yan, S., Cai, R., Clarke, C. R., Morris, C. E., Schaad, N. W., Schuenzel, E. L., Lacy, G. H., Sun, X., & Jones, J. B. (2010). PAMDB, a multilocus sequence typing and analysis database and website for plant-associated microbes. Phytopathology, 100, 208–215.
Amato, P., Parazols, M., Sancelme, M., Laj, P., Mailhot, G., & Delort, A.-M. (2006). Microorganisms isolated from the water phase of tropospheric clouds at the Puy de Dôme: Major groups and growth abilities at low temperatures. FEMS Microbiology Ecology, 59, 242–254.
Balestra, G., & Varvaro, L. (1997). Pseudomonas syringae pv. Syringae causal agent of disease on floral buds of Actinidia deliciosa (a. Chev) Liang et Ferguson in Italy. Journal of Phytopathology, 145, 375–378.
Balestra, G., Mazzaglia, A., & Rossetti, A. (2008a). Outbreak of bacterial blossom blight caused by Pseudomonas viridiflava on Actinidia chinensis kiwifruit plants in Italy. Plant Disease, 92, 1707–1707.
Balestra, G., Mazzaglia, A., Spinelli, R., Graziani, S., Quattrucci, A., & Rossetti, A. (2008b). Cancro batterico su Actinidia chinensis. L’Informatore Agrario, 38, 75–76.
Balestra, G. M., Perestrelo, L., Mazzaglia, A., & Rossetti, A. (2009). First report of blossom blight caused by pseudomonas syringae on kiwifruit plants in Portugal. Journal of Plant Pathology, 91, 231.
Bartoli, C., Berge, O., Monteil, C. L., Guilbaud, C., Balestra, G. M., Varvaro, L., Jones, C., Dangl, J. L., Baltrus, D. A., & Sands, D. C. (2014). The Pseudomonas viridiflava phylogroups in the P. syringae species complex are characterized by genetic variability and phenotypic plasticity of pathogenicity-related traits. Environmental Microbiology, 16, 2301–2315.
Bartoli, C., Lamichhane, J. R., Berge, O., Guilbaud, C., Varvaro, L., Balestra, G. M., Vinatzer, B. A., & Morris, C. E. (2015). A framework to gauge the epidemic potential of plant pathogens in environmental reservoirs: The example of kiwifruit canker. Molecular Plant Pathology, 16, 137–149. https://doi.org/10.1111/mpp.12167.
Berge, O., Monteil, C. L., Bartoli, C., Chandeysson, C., Guilbaud, C., Sands, D. C., & Morris, C. E. (2014). A user's guide to a data base of the diversity of Pseudomonas syringae and its application to classifying strains in this phylogenetic complex. PLoS One, 9, e105547.
Borschinger, B., Bartoli, C., Chandeysson, C., Guilbaud, C., Parisi, L., Bourgeay, J. F., Buisson, E., & Morris, C. E. (2016). A set of PCRs for rapid identification and characterization of Pseudomonas syringae phylogroups. Journal of Applied Microbiology, 120, 714–723. https://doi.org/10.1111/jam.13017.
Bultreys, A., & Kaluzna, M. (2010). Bacterial cankers caused by Pseudomonas syringae on stone fruit species with special emphasis on the Pathovars Syringae and Morsprunorum race 1 and race 2. Journal of Plant Pathology, 92, S1.
Chapman, J. R., Taylor, R. K., Weir, B. S., Romberg, M. K., Vanneste, J. L., Luck, J., & Alexander, B. J. R. (2012). Phylogenetic relationships among global populations of Pseudomonas syringae pv. actinidiae. Phytopathology, 102, 1034–1044. https://doi.org/10.1094/phyto-03-12-0064-r.
Crosse, J. (1963). Bacterial canker of stone-fruits. Annals of Applied Biology, 52, 97–104.
Crosse, J. (1966). Epidemiological relations of the pseudomonad pathogens of deciduous fruit trees. Annual Review of Phytopathology, 4, 291–310.
Cunty, A., Poliakoff, F., Rivoal, C., Cesbron, S., Fischer-Le, S. M., Lemaire, C., Jacques, M. A., Manceau, C., & Vanneste, J. L. (2015). Characterization of Pseudomonas syringae pv. actinidiae (Psa) isolated from France and assignment of Psa biovar 4 to a de novo pathovar: Pseudomonas syringae pv. actinidifoliorum pv. Nov. Plant Pathology, 64, 582–596. https://doi.org/10.1111/ppa.12297.
Drummond A. J., Ashton B., Buxton S., Cheung M., Cooper A., Heled J. & Kearse M. (2013). Geneious v6.1 created by Biomatters. Retrieved 27 January, 2013, from http://www.geneious.com
Dye, D., Bradbury, J., Goto, M., Hayward, A., Lelliott, R., & Schroth, M. (1980). International standards for naming pathovars of phytopathogenic bacteria and a list of pathovar names and pathotype strains. Review of Plant Pathology, 59, 153–168.
Edgar, R. C. (2004). MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Research, 32, 1792–1797.
Everett K. R. & Henshall W. R (1994). Epidemiology and population ecology of kiwifruit blossom blight. Plant Pathology, 43, 824–830.
Everett K. R., Taylor R. K., Romberg M. K., Rees-George J., Fullerton R. A., Vanneste J. L. & Manning M. A. (2011). First report of Pseudomonas syringae pv. actinidiae causing kiwifruit bacterial canker in New Zealand. Australasian Plant Disease Notes, 6, 67-71. https://doi.org/10.1007/s13314-011-0023-9.
Ferrante, P., & Scortichini, M. (2009). Identification of pseudomonas syringae pv. Actinidiae as causal agent of bacterial canker of yellow kiwifruit (Actinidia chinensis Planchon) in Central Italy. Journal of Phytopathology, 157, 768–770.
Frampton R. A., Taylor C., Holguín Moreno A. V., Visnovsky S. B., Petty N. K., Pitman A. R. & Fineran P. C. (2014). Identification of bacteriophages for the biocontrol of the kiwifruit canker phytopathogen Pseudomonas syringae pv. actinidiae. Applied and Environmental Microbiology, 80(7), 2216 https://doi.org/10.1128/aem.00062-14.
Fujikawa, T., & Sawada, H. (2016). Genome analysis of the kiwifruit canker pathogen pseudomonas syringae pv. Actinidiae biovar 5. Scientific Reports, 6, 21399.
Fujikawa, T., & Sawada, H. (2019). Genome analysis of pseudomonas syringae pv. Actinidiae biovar 6, which produces the phytotoxins, phaseolotoxin and coronatine. Scientific Reports, 9. https://doi.org/10.1038/s41598-019-40754-9.
Gardan, L., Shafik, H., Belouin, S., Broch, R., Grimont, F., & Grimont, P. (1999). DNA relatedness among the pathovars of Pseudomonas syringae and description of Pseudomonas tremae sp. nov. and Pseudomonas cannabina sp. nov.(ex Sutic and Dowson 1959). International Journal of Systematic Bacteriology, 49, 469–478.
Garlant, L., Koskinen, P., Rouhiainen, L., Laine, P., Paulin, L., Auvinen, P., Holm, L., & Pirhonen, M. (2013). Genome sequence of Dickeya solani, a new soft rot pathogen of potato, suggests its emergence may be related to a novel combination of non-ribosomal peptide/polyketide synthetase clusters. Diversity, 5, 824–842.
Glaeser, S. P., & Kämpfer, P. (2015). Multilocus sequence analysis (MLSA) in prokaryotic taxonomy. Systematic and Applied Microbiology, 38, 237–245. https://doi.org/10.1016/j.syapm.2015.03.007.
Gomila, M., Peña, A., Mulet, M., Lalucat, J., & García-Valdés, E. (2015). Phylogenomics and systematics in pseudomonas. Frontiers in Microbiology, 6. https://doi.org/10.3389/fmicb.2015.00214.
González, A. J., Rodicio, M. R., & Mendoza, M. C. (2003). Identification of an emergent and atypical Pseudomonas viridiflava lineage causing bacteriosis in plants of agronomic importance in a Spanish region. Applied and Environmental Microbiology, 69, 2936–2941.
Goode, M. J., & Sasser, M. (1980). Prevention-the key to controlling bacterial spot and bacterial speck of tomato. Plant Disease, 64, 831–834.
Guilbaud, C., Morris, C. E., Barakat, M., Ortet, P., & Berge, O. (2016). Isolation and identification of Pseudomonas syringae facilitated by a PCR targeting the whole P. syringae group. FEMS Microbiology Ecology, 92(1).
Guindon, S., Dufayard, J.-F., Lefort, V., Anisimova, M., Hordijk, W., & Gascuel, O. (2010). New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Systematic Biology, 59, 307–321. https://doi.org/10.1093/sysbio/syq010.
Harper, S., Ward, L., & Clover, G. (2010). Development of LAMP and real-time PCR methods for the rapid detection of Xylella fastidiosa for quarantine and field applications. Phytopathology, 100, 1282–1288.
Hirano, S. S., & Upper, C. D. (1990). Population biology and epidemiology of Pseudomonas syringae. Annual Review of Phytopathology, 28, 155–177.
Hu, F., Fang, D., Young, J., & Xie, L. (1998). Identification of the pathogen caused bacterial blight of kiwifruit in China. Acta Phytopathologica Sinica, 28, 175–181.
Hu, F. P., Young, J. M., & Jones, D. S. (1999). Evidence that bacterial blight of kiwifruit, caused by a Pseudomonas sp., was introduced into New Zealand from China. Journal of Phytopathology, 147, 89–97.
Hwang, M. S., Morgan, R. L., Sarkar, S. F., Wang, P. W., & Guttman, D. S. (2005). Phylogenetic characterization of virulence and resistance phenotypes of Pseudomonas syringae. Applied and Environmental Microbiology, 71, 5182–5191.
Ieki H. (1993). Kiwifruit diseases in Japan. Japan Pesticide Information, 11–13.
Jakob, K., Goss, E. M., Araki, H., Van, T., Kreitman, M., & Bergelson, J. (2002). Pseudomonas viridiflava and P. syringae - natural pathogens of Arabidopsis thaliana. Molecular Plant-Microbe Interactions, 15, 1195–1203. https://doi.org/10.1094/mpmi.2002.15.12.1195.
Kałużna, M., Willems, A., Pothier, J. F., Ruinelli, M., Sobiczewski, P., & Puławska, J. (2016). Pseudomonas cerasi sp. nov.(non griffin, 1911) isolated from diseased tissue of cherry. Systematic and Applied Microbiology, 39, 370–377.
Koh, Y. J., Lee, D. H., Shin, J. S., & Hur, J.-S. (2001). Chemical and cultural control of bacterial blossom blight of kiwifruit caused by Pseudomonas syringae in Korea. New Zealand Journal of Crop and Horticultural Science, 29, 29–34.
Lamichhane, J. R., Varvaro, L., Parisi, L., Audergon, J.-M., & Morris, C. E. (2014). Disease and frost damage of woody plants caused by Pseudomonas syringae: Seeing the forest for the trees. Advances in Agronomy, 126, 235–295.
Lee, Y. S., Han, H. S., Kim, G. H., Koh, Y. J., Hur, J.-S., & Jung, J. S. (2009). Causal agents of blossom blight of kiwifruit in Korea. Plant Pathology Journal, 25, 220–224. https://doi.org/10.5423/ppj.2009.25.3.220.
Lelliott, R., Billing, E., & Hayward, A. (1966). A determinative scheme for the fluorescent plant pathogenic pseudomonads. Journal of Applied Microbiology, 29, 470–489.
Lindemann, J., Arny, D. C., & Upper, C. D. (1984). Use of an apparent infection threshold population of Pseudomonas syringae to predict incidence and severity of brown spot of bean. Phytopathology, 74, 1334–1339.
Loper, J. E., Hassan, K. A., Mavrodi, D. V., Davis, E. W., II, L. C. K., Shaffer, B. T., Elbourne, L. D. H., Stockwell, V. O., Hartney, S. L., Breakwell, K., Henkels, M. D., Tetu, S. G., Rangel, L. I., Kidarsa, T. A., Wilson, N. L., van de Mortel, J. E., Song, C., Blumhagen, R., Radune, D., Hostetler, J. B., Brinkac, L. M., Durkin, A. S., Kluepfel, D. A., Wechter, W. P., Anderson, A. J., Kim, Y. C., Pierson, L. S., III, P. E. A., Lindow, S. E., Kobayashi, D. Y., Raaijmakers, J. M., Weller, D. M., Thomashow, L. S., Allen, A. E., & Paulsen, I. T. (2012). Comparative Genomics of Plant-Associated Pseudomonas spp.: Insights into Diversity and Inheritance of Traits Involved in Multitrophic Interactions. PLoS Genet, 8, e1002784. https://doi.org/10.1371/journal.pgen.1002784.
Lovell, H. C., Mansfield, J. W., Godfrey, S. A., Jackson, R. W., Hancock, J. T., & Arnold, D. L. (2009). Bacterial evolution by genomic island transfer occurs via DNA transformation in planta. Current Biology, 19, 1586–1590.
Maiden, M. C., Bygraves, J. A., Feil, E., Morelli, G., Russell, J. E., Urwin, R., Zhang, Q., Zhou, J., Zurth, K., & Caugant, D. A. (1998). Multilocus sequence typing: A portable approach to the identification of clones within populations of pathogenic microorganisms. Proceedings of the National Academy of Sciences, 95, 3140–3145.
Marchi, G., Viti, C., Giovannetti, L., & Surico, G. (2005). Spread of Levan-positive populations of pseudomonas savastanoi pv. Savastanoi, the causal agent of olive knot, in Central Italy. European Journal of Plant Pathology, 112, 101–112. https://doi.org/10.1007/s10658-005-0804-0.
Mazzaglia, A., Studholme, D. J., Taratufolo, M. C., Cai, R., Almeida, N. F., Goodman, T., Guttman, D. S., Vinatzer, B. A., & Balestra, G. M. (2012). Pseudomonas syringae pv. actinidiae (PSA) isolates from recent bacterial canker of kiwifruit outbreaks belong to the same genetic lineage. PLoS ONE, 7, e36518.
McCann, H. C., Rikkerink, E. H., Bertels, F., Fiers, M., Lu, A., Rees-George, J., Andersen, M. T., Gleave, A. P., Haubold, B., & Wohlers, M. W. (2013). Genomic analysis of the kiwifruit pathogen Pseudomonas syringae pv. actinidiae provides insight into the origins of an emergent plant disease. PLoS Pathog, 9, e1003503.
Miyoshi, T., & Tachibana, Y. (1996). Epidemiological studies on bacterial blossom blight of kiwifruit 1. Differences in susceptibility of flower bud to pseudomonas syringae, a causal pathogen of bacterial blossom blight. Annals of the Phytopathological Society of Japan, 62, 517–522.
Monteil, C. L., Guilbaud, C., Glaux, C., Lafolie, F., Soubeyrand, S., & Morris, C. E. (2012). Emigration of the plant pathogen Pseudomonas syringae from leaf litter contributes to its population dynamics in alpine snowpack. Environmental Microbiology, 14, 2099–2112. https://doi.org/10.1111/j.1462-2920.2011.02680.x.
Morán, F., Marco-Noales, E., Escrich, A., Barbé, S., & López, M. M. (2018). Biodiversity and biogeography of three pseudomonas syringae Pathovars which affect kiwi fruit cultivation. Biodiversity Online Journal, 1, 1–3.
Morris, C. E., Sands, D. C., Vinatzer, B. A., Glaux, C., Guilbaud, C., Buffiere, A., Yan, S., Dominguez, H., & Thompson, B. M. (2008). The life history of the plant pathogen Pseudomonas syringae is linked to the water cycle. The ISME Journal, 2, 321–334.
Morris, C. E., Bardin, M., Kinkel, L. L., Moury, B., Nicot, P. C., & Sands, D. C. (2009). Expanding the paradigms of plant pathogen life history and evolution of parasitic fitness beyond agricultural boundaries. PLoS Pathogens, 5, e1000693.
O'Brien, H. E., Thakur, S., Gong, Y., Fung, P., Zhang, J., Yuan, L., Wang, P. W., Yong, C., Scortichini, M., & Guttman, D. S. (2012). Extensive remodeling of the Pseudomonas syringae pv. avellanae type III secretome associated with two independent host shifts onto hazelnut. BMC Microbiology, 12. https://doi.org/10.1186/1471-2180-12-141.
Parkinson, N., Bryant, R., Bew, J., & Elphinstone, J. (2011). Rapid phylogenetic identification of members of the Pseudomonas syringae species complex using the rpoD locus. Plant Pathology, 60, 338–344. https://doi.org/10.1111/j.1365-3059.2010.02366.x.
Pitman, A. R., Jackson, R. W., Mansfield, J. W., Kaitell, V., Thwaites, R., & Arnold, D. L. (2005). Exposure to host resistance mechanisms drives evolution of bacterial virulence in plants. Current Biology, 15, 2230–2235.
Purahong, W., Orru, L., Donati, I., Perpetuini, G., Cellini, A., Lamontanara, A., Michelotti, V., Tacconi, G., & Spinelli, F. (2018). Plant microbiome and its link to plant health: Host species, organs and pseudomonas syringae pv. Actinidiae infection shaping bacterial Phyllosphere communities of kiwifruit plants. Frontiers in Plant Science, 9. https://doi.org/10.3389/fpls.2018.01563.
Rees-George, J., Vanneste, J. L., Cornish, D. A., Pushparajah, I. P. S., Yu, J., Templeton, M. D., & Everett, K. R. (2010). Detection of Pseudomonas syringae pv. actinidiae using polymerase chain reaction (PCR) primers based on the 16S–23S rDNA intertranscribed spacer region and comparison with PCR primers based on other gene regions. Plant Pathology, 59, 453–464. https://doi.org/10.1111/j.1365-3059.2010.02259.x.
Sarkar, S. F., & Guttman, D. S. (2004). Evolution of the Core genome of Pseudomonas syringae, a highly clonal, endemic plant pathogen. Applied and Environmental Microbiology, 70, 1999–2012. https://doi.org/10.1128/aem.70.4.1999-2012.2004.
Sarris P. F., Trantas E. A., Mpalantinaki E., Ververidis F. & Goumas D. E. (2012). Pseudomonas viridiflava, a multi host plant pathogen with significant genetic variation at the molecular level, Pseudomonas viridiflava, a Multi Host Plant Pathogen with Significant Genetic Variation at the Molecular Level.
Schober, B. M., & Zadoks, J. C. (1999). Survival of softrot bacteria during the storage of witloof chicory roots. Journal of Phytopathology-Phytopathologische Zeitschrift, 147, 461–466.
Scortichini, M., Marcelletti, S., Ferrante, P., Petriccione, M., & Firrao, G. (2012). Pseudomonas syringae pv. actinidiae: A re-emerging, multi-faceted, pandemic pathogen. Molecular Plant Pathology, 13, 631–640. https://doi.org/10.1111/j.1364-3703.2012.00788.x.
Shaffer, B. T., & Lighthart, B. (1997). Survey of culturable airborne bacteria at four diverse locations in Oregon: Urban, rural, forest, and coastal. Microbial Ecology, 34, 167–177.
Stockwell, V. O., Johnson, K. B., Sugar, D., & Loper, J. E. (2010). Control of fire blight by Pseudomonas fluorescens A506 and Pantoea vagans C9-1 applied as single strains and mixed Inocula. Phytopathology, 100, 1330–1339. https://doi.org/10.1094/phyto-03-10-0097.
Straub, C., Colombi, E., Li, L., Huang, H., Templeton, M. D., McCann, H. C., & Rainey, P. B. (2018). The ecological genetics of Pseudomonas syringae from kiwifruit leaves. Environmental Microbiology, 20, 2066–2084. https://doi.org/10.1111/1462-2920.14092.
Takikawa, Y., Serizawa, S., Ichikawa, T., Tsuyumu, S., & Goto, M. (1989). Pseudomonas syringae pv. actinidiae pv. Nov = the causal bacterium of canker of kiwifruit in Japan. Japanese Journal of Phytopathology, 55, 437–444. https://doi.org/10.3186/jjphytopath.55.437.
Tomlinson, J., & Boonham, N. (2008). Potential of LAMP for detection of plant pathogens. CAB reviews: Perspectives in agriculture, veterinary science. Nutrition and Natural Resources, 3, 1–7.
Tyson, J. L., Rees-George, J., Curtis, C. L., Manning, M. A., & Fullerton, R. A. (2012). Survival of Pseudomonas syringae pv. actinidiae on the orchard floor over winter. New Zealand Plant Protection, 65, 25–28.
Vanneste, J. L., Yu, J., Cornish, D. A., Tanner, D. J., Windner, R., Chapman, J. R., Taylor, R. K., Mackay, J. F., & Dowlut, S. (2013). Identification, virulence, and distribution of two biovars of Pseudomonas syringae pv. actinidiae in New Zealand. Plant Disease, 97, 708–719. https://doi.org/10.1094/pdis-07-12-0700-re.
Vicente, J. G., Alves, J. P., Russell, K., & Roberts, S. J. (2004). Identification and discrimination of Pseudomonas syringae isolates from wild cherry in England. European Journal of Plant Pathology, 110, 337–351.
Visnovsky, S. B., Fiers, M., Lu, A., Panda, P., Taylor, R., & Pitman, A. R. (2016). Draft genome sequences of 18 strains of Pseudomonas isolated from kiwifruit plants in New Zealand and overseas. Genome Announcements, 4. https://doi.org/10.1128/genomeA.00061-16.
Wilkie, J. P., Dye, D. W., & Watson, D. R. W. (1973). Further hosts of Pseudomonas viridiflava. New Zealand Journal of Agricultural Research, 16, 315–323.
Young, J. M. (1987). New plant disease record in New Zealand: Pseudomonas syringae pv. persicae from nectarine, peach, and Japanese plum. New Zealand Journal of Agricultural Research, 30, 235–247.
Young, J. M. (1988). Bacterial blight of kiwifruit in New Zealand. Bulletin OEPP, 18, 131–140.
Young, J. M. (2010). Taxonomy of Pseudomonas syringae. Journal of Plant Pathology, S5–S14.
Young J. M. & Fletcher M. J. (1997). International Collection of Micro-organisms from Plants: catalogue accessions 1-12989. (Landcare research, PO Box 40, Lincoln 8152, New Zealand.
Young, J. M., Gardan, L., Ren, X. Z., & Hu, F. P. (1997). Genomic and phenotypic characterization of the bacterium causing blight of kiwifruit in New Zealand. Plant Pathology, 46, 857–864.
Acknowledgements
This work was funded by the Better Border Biosecurity programme (B3) (www.b3nz.org).
The authors thanks Drs Erik Rikkerink, Hayley Ridgway and Faulk Kalamorz from The New Zealand Institute for Plant and Food Research Ltd. for their valuable feedback to the manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
The manuscript complies with ethical rules of the European Journal of Plant Pathology, as reported in the “Ethical Responsibilities of Authors” in the “Instruction for Authors” section.
Conflict of interest and human or animal participants
There are no potential conflicts of interest, and the research does not involve human participants and / or animals.
Ethical approval
All authors have approved the manuscript and agreed with its submission to European Journal of Plant Pathology.
Electronic supplementary material
Figure S1
An unrooted Maximum Likelihood tree (Guindon et al. 2010) constructed using concatenated partial DNA sequences of gyrB and gapA from pseudomonads collected from kiwifruit plants, other hosts and the environment. The pseudomonads included P. viridiflava strains and other pseudomonads downloaded from the Plant Associated and Environmental Microbes Database (PAMDB). Twenty-seven isolates Group A from our collection from kiwifruit and from Prunus were also included. This analysis supported our multilocus sequence analysis (MLSA) data, confirming the identity of ABAC 43 and ICMP 11289 as P. viridiflava. ICMP 3272 and ICMP 13303, which were considered P. viridiflava, did not cluster with the P. viridiflava isolates, indicating that they are probably P. syringae phylogroup III. (PNG 449 kb)
Table S1
(DOCX 20 kb)
Table S2
(DOCX 15.9 kb)
Rights and permissions
About this article
Cite this article
Visnovsky, S.B., Marroni, M.V., Pushparajah, S. et al. Using multilocus sequence analysis to distinguish pathogenic from saprotrophic strains of Pseudomonas from stone fruit and kiwifruit. Eur J Plant Pathol 155, 643–658 (2019). https://doi.org/10.1007/s10658-019-01799-8
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10658-019-01799-8