Abstract
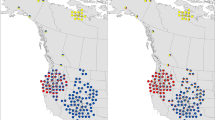
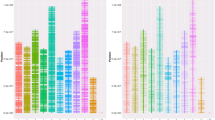
Bald eagles (Haliaeetus leucocephalus) underwent a severe population bottleneck in the mid-1900s due to Dichlorodiphenyltrichloroethane (DDT) use as an insecticide. After its ban in 1972, the population began to recover with the increase being attributed to banning DDT and reintroduction/translocation programs. Although bald eagles have increased and may be in a phase of exponential growth, many populations continue to experience anthropogenic stressors. Assessing levels of standing genetic variation and the partitioning of genetic variation within and among populations is critical for the development of informed conservation management plans. To begin addressing these concerns, we developed a custom 50K Affymetrix Axiom myDesign single nucleotide polymorphism (SNP) array and performed preliminary population genomic analyses on geographically disparate populations of bald eagles to test the utility of this SNP array for assessing levels of standing genetic variation in the gene pool and determining the partitioning of genetic variation within and among populations. To develop the array, a combination of RAD-tag sequencing and genome sequencing was used with the final chip consisting of 50,789 SNPs in both genic and intergenic regions of the genome. After genotyping 169 hatchlings, 45,952 SNPs from the array were found to be of quality and were used in Structure, Admixture, and a discriminant analysis of principal components (DAPC) analyses. Results from all of these analyses indicate that despite the significant population bottleneck, sufficient genetic variation is detectable within the bald eagle gene pool. Moreover, based on our disparate geographic sampling of bald eagles, our preliminary analyses indicate statistically significant partitioning of the genetic variation among broad sampling areas.



Similar content being viewed by others
References
Alexander DH, Novembre J, Lange K (2009) Fast model-based estimation of ancestry in unrelated individuals. Genome Res 19:1655–1664
Alison LJ, Driscoll JT, Jacobson KV (2008) Demographic analysis of the bald eagle in Arizona. Nongame and Endangered Wildlife Program Technical Report 221. Arizona Game and Fish Department, Phoenix, Arizona
Arnold B, Corbett-Detig RB, Hartl D, Bomblies K (2013) RADseq underestimates diversity and introduces genealogical biases due to nonrandom haplotype sampling. Mol Ecol 22:3179–3190
Beatty GL, Driscoll JT (1996) Identity of breeding bald eagles in Arizona 1991–1995. Nongame and Endangered Wildlife Program Technical Report 92. Arizona Game and Fish Department, Phoenix, Arizona
Benesten L, Gosselin T, Perrier C, Sainte-Marie B, Rochette R, Bernatchex L (2015) RAD genotyping reveals fine-scale genetic structuring and provides powerful population assignment in a widely distributed marine species, the American lobster (Homarus americanus). Mol Ecol 24:299–315
Blanco-Bercial L, Bucklin A (2016) New view of population genetics of zooplankton: RAD-seq analysis reveals population structure of the North Atlantic planktonic copepod Centropages typicus. Mol Ecol 25:1566–1580
Bourret V, Kent MP, Primmer CR, Vasemagi A, Karlsson S, Hindar K et al (2013) SNP-array reveals genome-wide patterns of geographical and potential adaptive divergence across the natural range of Atlantic salmon (Salmo salar). Mol Ecol 22:532–551
Bowerman WW, Giesy JP, Best DA, Kramer VJ (1995) A review of factors affecting productivity of bald eagles in the great lakes region: implications for recover. Environ Health Perspect 103:51–59
Bristol RM, Tucker R, Dawson DA, Horsburgh G, Prys-Jones RP, Frantz AC, Krupa A et al (2013) Comparison of historical bottleneck effects and genetic consequences of re-introduction in a critically endangered island passerine. Mol Ecol 22:4644–4662
Brown JW, Van Coeverdeen De Groot PJ, Birt TP, Seutin G, Boag PT, Friesen VL (2007) Appraisal of the consequences of the DDT-induced bottleneck on the level and geographic distribution of neutral genetic variation in Canadian peregrine falcons, Falco peregrinus. Mol Ecol 16:327–343
Cammen KM, Schultz TF, Rosel PE, Wells RS, Read AJ (2015) Genome wide investigation of adaptation to harmful algal blooms in common bottlenose dolphins (Tursiops truncates). Mol Ecol 24:4697–4710
Center for Biological Diversity (2007) [Internet]. Memos: Bureaucrats Overrule Scientists on Desert Nesting Bald Eagle Delisting. http://www.biologicaldiversity.org/swcbd/press/desert-bald-eagle-05-17-2007.html. Accessed 1 Aug 2017
Cingolani P, Platts A, Wang LL et al (2012) A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly 6:1–13
Danecek P, Auton A, Abecasis G, Albers CA, Banks E, DePristo MA et al (2011) The variant call format and VCFtools. Bioinformatics 27:2156–2158
Environmental Alaska (2014) [Internet]. Bald eagles in Alaska. http://environmentalaska.us/bald-eagles.html. Accessed 1 Aug 2017
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14:2611–2620
Ewing SR, Nager RG, Nicoll MAC, Aumjaud A, Jones CG, Keller LF (2008) Inbreeding and loss of genetic variation in a reintroduced population of Mauritius Kestrel. Conserv Biol 22:395–404
Excoffier L, Lischer HEL (2010) Arlequin suite ver 3.5: a new series of programs to perform population genetic analysis under Linux and Windows. Mol Ecol Res 10:564–567
Georgia Department of Natural Resources (2017) [Internet]. The bald eagle in Georgia. http://georgiawildlife.com/bald-eagle. Accessed 1 Aug 2017
Godoy JA, Negro JJ, Hiraldo F, Donazar JA (2004) Phylogeography, genetic structure and diversity in the endangered bearded vulture as revealed by mitochondrial DNA. Mol Ecol 13:371–390
Hess JE, Campbell NR, Close DA, Docker MF, Narum SR (2013) Population genomics of Pacific lamprey: adaptive variation in a highly dispersive species. Mol Ecol 22:2898–2916
Hoffberg SL, Kieran TJ, Catchen JM, Devault A, Faircloth BC, Mauricio R, Glenn TC (2016) RADcap: sequence capture of dual-digest RADseq libraries with identifiable duplicates and reduced missing data. Mol Ecol Resour 16:1264–1278
Hohenlohe PA, Bassham S, Etter PD, Stiffler N, Johnson EA, Cresko WA (2010) Population genomics of parallel adaptation in threespine stickleback using sequenced RAD Tags. PLoS Genet 6:1–23
Jombart T (2008) Adegenet: a R package for the multivariate analysis of genetic markers. Bioinformatics 24:1403–1405
Kjeldsen S, Zenger KR, Leigh K, Ellis W, Tobey J, Phalen D et al (2016) Genome-wide SNP loci reveal novel insights into koala (Phascolarctos cinereus) population variability across its range. Conserv Genet 17:337–353
Langmead B, Salzberg S (2013) Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–359
Latch EK, Dharmarajan G, Glaubitz JS, Rhodes OE (2006) Relative performance of Bayesian clustering software for inferring population substructure and individual assignment at low levels of population differentiation. Conserv Genet 7:295–302
Li H (2011) A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequence data. Bioinformatics 27:2987–2993
Lish JW, Sherrod SK (2007) A history of bald eagle nesting activity in Oklahoma. Proc Okla Acad Sci 20:15–20
Longmire JL, Maltbie M, Baker RJ (1997) Use of “lysis buffer” in DNA isolation and its implication for museum collections. Occas Pap Mus Texas Tech Univ 163:1–3
Malenfant RM, Coltman DW, Davis CS (2015) Design of a 9 K illumine BeadChip for polar bears (Ursus meritimus) from RAD and transcriptome sequencing. Mol Ecol Resour 15:587–600
Martinez-Cruz B, Godoy JA, Negro JJ (2004) Population genetics after fragmentation: the case of the endangered Spanish imperial eagle. Mol Ecol 13:2243–2255
Maruyama T, Fuerst PA (1985) Population bottlenecks and nonequilibrium models in population genetics. II. Number of alleles in a small population that was formed by a recent bottleneck. Genetics 111:675–689
Jacobson KV, McCarty, KM, Driscoll JT (2006) Arizona bald eagle management program 2006 summary report. Nongame and Endangered Wildlife Program Technical Report 239. Arizona Game and Fish Department, Phoenix, Arizona
McEwan L, Hirth DH (2012) Southern bald eagle productivity and nest site selection. J Wildl Manag 43:585–594
Millsap B, Breen T, McConnell E, Steffer T, Phillips L, Douglass N et al (2004) Comparative fecundity and survival of bald eagles fledged from suburban and rural natal areas in Florida. J Wildl Manag 68:1018–1031
Millsap BA, Harmata AR, Stahlecker DW, Mikesic DG (2014) Natal dispersal distance of bald and golden eagles originating in the coterminous United States as inferred from band encounters. J Raptor Res 48:13–23
Mojica EK, Watts BD, Turrin CL (2016) Utilization probability map for migrating bald eagles in northeastern North America: a tool for siting wind energy facilities and other flight hazards. PLoS ONE 11:1–11
Nei M, Maruyama T, Chakraborty R (1975) The bottleneck effect and genetic variability in populations. Evolution 29:1–10
Newton I (1979) Population ecology of raptors. Buteo Books, Vermillion
North Carolina Wildlife Resources Commission (2005) [Internet]. Bald Eagle Fact Sheet; http://www.ncwildlife.org/pg07_wildlifespeciescon/nongame_baldeagle_hires.pdf. Accessed 1 Aug 2017
Ouborg NJ, Pertoldi C, Loeschocke V, Bijlsma R, Hedrick PW (2010) Conservation genetics in transition to conservation genomics. Trends Genet 26:177–187
Pagel JE, Kritz KJ, Millsap BA, Murphy RK, Kershner EL, Covington S (2013) Bald eagle and golden eagle mortalities at wind energy facilities in the contiguous United States. J Raptor Res 47:311–315
Pilot M, Greco C, vonHoldt BM, Jedrzejewska B, Randi E, Jedrzejewski W, Sidorovich VE et al (2014) Genome-wide signatures of population bottlenecks and diversifying selection in European wolves. Heredity 112:428–442
Poulakakis N, Antoniou A, Mantziou G, Parmakelis A, Skartsi T, Vasilakis D, Elorriaga J, De La Puente J et al (2008) Population structure, diversity, and phylogeography in the near-threatened Eurasian black vultures. Nat Hist 95:859–872
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Pujolar JM, Jacobsen MW, Frydenberg J, Larsen PF, Maes GE, Zane L et al (2013) A resource of genome-wide single-nucleotide polymorphisms generated by RAD tag sequencing in the critically endangered European eel. Mol Ecol Resour 13:706–714
Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MAR, Bender D et al (2007) Report Plink: a tool set for whole-genome association and population-based linkage analysis. Am J Hum Genet 81:559–575
Raymond M, Rousset F (1995) Genepop (version 1.2) Population genetics software for exact tests and ecumenicism. J Hered 86:248–249
Ruiz-Gonzalez A, Cushman SA, Madeira JJ, Randi E, Gomez-Moliner BJ (2015) Isolation by distance, resistance and/or clusters? Lessons learned from a forest-dwelling carnivore inhabiting a heterogeneous landscape. Mol Ecol 24:5110–5129
Seddon JM, Parker HG, Ostrander EA, Ellegren H (2005) SNPs in ecological and conservation studies: a test in the Scandinavian wolf population. Mol Ecol 14:503–511
Simons T, Sherrod SK, Collopy MW, Jenkins MA (1988) Restoring the bald eagle. Am Sci 76:253–260
Sprunt A, Robertson WB, Postupalsky S, Hensel RJ, Knoder CF, Ligas FJ (1973) Comparative productivity of six bald eagle populations. Trans N Am Wildl Conf 38:96–100
Stauber E, Finch N, Talcott PA, Gray JM (2010) Lead poisoning of bald (Haliaeetus leucocephalus) and golden (Aquila chrysaetos) eagles in the U.S. inland Pacific northwest region—an 18 year retrospective study: 1991–2008. J Avian Med Surg 24:279–298
Stinchcombe JR, Hoekstra HE (2008) Combining population genomics and quantitative genetics: finding the genes underlying ecologically important traits. Heredity 100:158–170
van Bers NEM, Santure AW, van Oers K, Cauwer ID, Dibbits BW, Mateman C et al (2016) The design and cross-population application of a genome-wide SNP chip for the great tit, Parus major. Mol Ecol Resour 12:753–770
Ventana Wildlife Society (2012) [Internet]. Bald eagle recovery. http://www.ventanaws.org/species_eagles/. Accessed 1 Aug 2017
Watts BD, Therres GD, Byrd MA (2008) Recovery of the chesapeake bay bald eagle nesting population. J Wildl Manag 72:152–158
Acknowledgements
We would like to thank Bryan Bedrosian, Brian Millsap, Erica Miller, Kenneth (“Tuk”) Jacobson, Russ Horton, Todd Katzner, Victor Roubidoux, and Victoria Vosberg for contributing the bald eagle blood samples used in this study. Without their generosity this study could not have been completed. We would also like to thank the Iowa Tribe of Oklahoma, Shakopee Mdewakanton Sioux, and the Eppley Foundation for providing the funding necessary for this project. Thanks are also extended to the Ford Foundation for funding Megan Judkins research fellowship while the project was being conducted. This manuscript represents one of four chapters that were part of Megan’s dissertation and therefore a special thanks to the other members of her dissertation committee, Drs. Meredith Hamilton, Andrew Doust, and Jim Lish. Their efforts on previous versions of this manuscript were invaluable.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Judkins, M.E., Couger, B.M., Warren, W.C. et al. A 50K SNP array reveals genetic structure for bald eagles (Haliaeetus leucocephalus). Conserv Genet 21, 65–76 (2020). https://doi.org/10.1007/s10592-019-01216-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10592-019-01216-x