Abstract
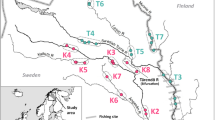
The restoration and maintenance of habitat connectivity are major challenges in conservation biology. These aims are especially critical for migratory species using corridors that can be obstructed by anthropogenic barriers. Here, we explored the origins and genetic diversity of Atlantic salmon (Salmo salar) recolonizing upstream areas of the largest South European Atlantic salmon population (Adour drainage, France) following restoration of connectivity and stocking. We genotyped 1,009 juvenile individuals, sampled either in continuously inhabited downstream sites or in recently reconnected and recolonized upstream locations, at 12 microsatellite loci. We found significant fine scale genetic structure, with three main genetic clusters corresponding to the Nive, Nivelle and Gaves rivers. Within each of these clusters, samples collected in continuously inhabited and recently recolonized sites had comparable allelic richness and effective population sizes and were only weakly differentiated. Genetic structure among basins was also similar among continuously inhabited and recently recolonized sites. The majority of the individuals sampled from recently recolonized sites were assigned to neighboring continuously inhabited downstream sites, but noticeable proportions of fish were assigned to samples collected in more distant sites or identified as putative hybrids. Overall, this study suggests that the restoration of accessibility to upstream areas can allow for the recolonization and effective reproduction of Atlantic salmon from proximate downstream refugia, which does not decrease local diversity or disrupt existing genetic structure.





Similar content being viewed by others
References
Aprahamian MW, Smith KM, McGinnity P, McKelvey S, Taylor J (2003) Restocking of salmonids—opportunities and limitations. Fish Res 62:211–227
Araki H, Schmid C (2010) Is hatchery stocking a help or harm? Evidence, limitations and future directions in ecological and genetic surveys. Aquaculture 308:S2–S11. doi:10.1016/j.aquaculture.2010.05.036
Araki H, Ardren WR, Olsen E, Cooper B, Blouin MS (2007) Reproductive success of captive-bred steelhead trout in the wild: evaluation of three hatchery programs in the Hood River. Conserv Biol 21(1):181–190. doi:10.1111/j.1523-1739.2006.00564.x
Araki H, Berejikian BA, Ford MJ, Blouin MS (2008) Fitness of hatchery-reared salmonids in the wild. Evol Appl 1(2):342–355. doi:10.1111/j.1752-4571.2008.00026.x
Aronson J (2011) Ecological restoration in 2010: insight from four continents. Conserv Biol 25(1):206–208. doi:10.1111/j.1523-1739.2010.01621.x
Barracou D (2008) Synthèse de la qualité des frayères à saumons sur le bassin de l’Adour: accessibilité des géniteurs et survie des juvéniles. Atlantic Salmon Arc Project, Migradour
Baudouin L, Lebrun P (2000) An operational bayesian approach for the identification of sexually reproduced cross-fertilized populations using molecular markers. In: Dore C, Dosba F, Baril C (eds) International symposium on molecular markers for characterizing genotypes and identifying cultivars in horticulture. Montpellier, France, Mar 06–08 2000, pp 81–93
Beall E, Davaine P, Bazin D, Bousquet B (1995) Repeuplement du Gave de Pau. Utilisation de souches étrangères et locales domestiquées pour le repeuplement des rivières à Salmonidés. Rapport DIREN Midi Pyrénées code INRA 2713A, Station d’Hydrobiologie INRA, St Pée sur Nivelle
Beaudou D, Baril D, Roche B, LeBaron M, CattaneoBerrebi G, Berrebi P (1994) Recolonization in a devastated Corsican river: respective contribution of wild and domestic brown trout. In: International symposium on fish and their habitat. Lyon, France, Dec 06–08 1994, pp 259–266
Bednarek AT (2001) Undamming rivers: a review of the ecological impacts of dam removal. Environ Manag 27(6):803–814. doi:10.1007/s002670010189
Blanchet S, Rey O, Etienne R, Lek S, Loot G (2010) Species-specific responses to landscape fragmentation: implications for management strategies. Evol Appl 3(3):291–304. doi:10.1111/j.1752-4571.2009.00110.x
Boet P, Belliard J, Berrebi-dit-Thomas R, Tales E (1999) Multiple human impacts by the City of Paris on fish communities in the Seine river basin, France. Hydrobiologia 410:59–68
Bourret V, O’Reilly PT, Carr JW, Berg PR, Bernatchez L (2011) Temporal change in genetic integrity suggests loss of local adaptation in a wild Atlantic salmon (Salmo salar) population following introgression by farmed escapees. Heredity 106:500–510
Bourret V, Dionne M, Kent MP, Lien S, Bernatchez L (2013) Landscape Genomics in Atlantic Salmon (Salmo salar): searching for gene-environment interactions driving local adaptation. Evolution 67(12):3469–3487. doi:10.1111/evo.12139
Boylan P, Adams CE (2006) The influence of broad scale climatic phenomena on long term trends in Atlantic salmon population size: an example from the River Foyle, Ireland. J Fish Biol 68(1):276–283. doi:10.1111/j.0022-1112.2006.00893.x
Brown JJ, Limburg KE, Waldman JR, Stephenson K, Glenn EP, Juanes F, Jordaan A (2013) Fish and hydropower on the U.S. Atlantic coast: failed fisheries policies from half-way technologies. Conserv Lett. doi:10.1111/conl.12000
Charlesworth B (2009) Fundamental concepts in genetics: effective population size and patterns of molecular evolution and variation. Nat Rev Genet 10(3):195–205. doi:10.1038/nrg2526
Clewell AF, Aronson J (2006) Motivations for the restoration of ecosystems. Conserv Biol 20(2):420–428. doi:10.1111/j.1523-1739.2006.00340.x
Corander J, Waldmann P, Marttinen P, Sillanpaa MJ (2004) BAPS 2: enhanced possibilities for the analysis of genetic population structure. Bioinformatics 20(15):2363–2369. doi:10.1093/bioinformatics/bth250
Coulon A, Fitzpatrick JW, Bowman R, Lovette IJ (2010) Effects of habitat fragmentation on effective dispersal of Florida scrub-jays. Conserv Biol 24(4):1080–1088. doi:10.1111/j.1523-1739.2009.01438.x
Coutant CC, Whitney RR (2000) Fish behavior in relation to passage through hydropower turbines: a review. Trans Am Fish Soc 129(2):351–380
Couvet D (2002) Deleterious effects of restricted gene flow in fragmented populations. Conserv Biol 16(2):369–376. doi:10.1046/j.1523-1739.2002.99518.x
Davaine P, Beall E, Glise S (1996) Repeuplement du Gave de Pau : utilisation de souches domestiquées de saumon atlantique. INRA, St Pée sur Nivelle
De Groot RS, Blignaut J, Van Der Ploeg S, Aronson J, Elmqvist T, Farley J (2013) Benefits of investing in ecosystem restoration. Conserv Biol. doi:10.1111/cobi.12158
Dillane E, McGinnity P, Coughlan JP, Cross MC, de Eyto E, Kenchington E, Prodohl P, Cross TF (2008) Demographics and landscape features determine intrariver population structure in Atlantic salmon (Salmo salar L.): the case of the River Moy in Ireland. Mol Ecol 17:4786–4800
Dionne M, Caron F, Dodson J, Bernatchez L (2008) Landscape genetics and hierarchical genetic structure in Atlantic salmon: the interaction of gene flow and local adaptation. Mol Ecol 17(10):2382–2396
Dionne M, Caron F, Dodson JJ, Bernatchez L (2009) Comparative survey of within-river genetic structure in Atlantic salmon; relevance for management and conservation. Conserv Genet 10(4):869–879. doi:10.1007/s10592-008-9647-5
Dixo M, Metzger JP, Morgante JS, Zamudio KR (2009) Habitat fragmentation reduces genetic diversity and connectivity among toad populations in the Brazilian Atlantic Coastal Forest. Biol Conserv 142(8):1560–1569. doi:10.1016/j.biocon.2008.11.016
Do C, Waples RS, Peel D, Macbeth GM, Tillett BJ, Ovenden JR (2013) NeEstimator v2: re-implementation of software for the estimation of contemporary effective population size (Ne) from genetic data. Mol Ecol Resour. doi:10.1111/1755-0998.12157
Dumas J, Prouzet P (2003) Variability of demographic parameters and population dynamics of Atlantic salmon (Salmo salar L.) in a southwest French river. ICES J Mar Sci 60(2):356–370
Earl D, vonHoldt B (2012) STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv Genet Resour 4(2):359–361. doi:10.1007/s12686-011-9548-7
Eldridge WH, Naish KA (2007) Long-term effects of translocation and release numbers on fine-scale population structure among coho salmon (Oncorhynchus kisutch). Mol Ecol 16(12):2407–2421. doi:10.1111/j.1365-294X.2007.03271.x
Eldridge WH, Myers JM, Naish KA (2009) Long-term changes in the fine-scale population structure of coho salmon populations (Oncorhynchus kisutch) subject to extensive supportive breeding. Heredity 103(4):299–309. doi:10.1038/hdy.2009.69
Ellis JS, Sumner KJ, Griffiths AM, Bright DI, Stevens JR (2011) Population genetic structure of Atlantic salmon, Salmo salar L., in the River Tamar, southwest England. Fish Manag Ecol 18(3):233–245. doi:10.1111/j.1365-2400.2010.00776.x
Ensing D, Prodöhl PA, McGinnity P, Boylan P, O’Maoiléidigh N, Crozier WW (2011) Complex pattern of genetic structuring in the Atlantic salmon (Salmo salar L.) of the River Foyle system in northwest Ireland: disentangling the evolutionary signal from population stochasticity. Ecol Evol 1(3):359–372. doi:10.1002/ece3.32
Evanno G, Regnault S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation. Mol Ecol 14:2611–2620
Ewers RM, Didham RK (2006) Confounding factors in the detection of species responses to habitat fragmentation. Biol Rev 81(1):117–142. doi:10.1017/s1464793105006949
Excoffier L, Lischer HEL (2010) Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Resour 10(3):564–567. doi:10.1111/j.1755-0998.2010.02847.x
Fahrig L (2003) Effects of Habitat fragmentation on biodiversity. Annu Rev Ecol Evol Syst 34: 487–515. doi:10.2307/30033784
Finnengan AK, Stevens JR (2008) Assessing the long-term genetic impact of historical stocking events on contemporary populations of Atlantic salmon, Salmo salar. Fish Manag Ecol 15(4):315–326. doi:10.1111/j.1365-2400.2008.00616.x
Fleming IA (1996) Reproductive strategies of Atlantic salmon: ecology and evolution. Rev Fish Biol Fish 6(4):379–416. doi:10.1007/Bf00164323
Frankham R (2002) Introduction to conservation genetics. Cambridge University Press, Cambridge 617 pp
Frankham R (2005) Genetics and extinction. Biol Conserv 126(2):131–140. doi:10.1016/j.biocon.2005.05.002
Fraser DJ (2008) How well can captive breeding programs conserve biodiversity? A review of salmonids. Evol Appl 1(4):535–586. doi:10.1111/j.1752-4571.2008.00036.x
Fraser DJ, Bernatchez L (2001) Adaptive evolutionary conservation: towards a unified concept for defining conservation units. Mol Ecol 10(12):2741–2752. doi:10.1046/j.0962-1083.2001.01411.x
Fraser DJ, Jones MW, McParland TL, Hutchings JA (2007) Loss of historical immigration and the unsuccessful rehabilitation of extirpated salmon populations. Conserv Genet 8(3):527–546. doi:10.1007/s10592-006-9188-8
Garcia de Leaniz C, Fleming IA, Einum S, Verspoor E, Jordan WC, Consuegra S, Aubin-Horth N, Lajus D, Letcher BH, Youngson AF, Webb JH, Vollestad LA, Villanueva B, Ferguson A, Quinn TP (2007) A critical review of adaptive genetic variation in Atlantic salmon: implications for conservation. Biol Rev 82(2):173–211
Garcia-Vazquez E, Moran P, Martinez JL, Perez J, de Gaudemar B, Beall E (2000) Alternative mating strategies in Atlantic salmon and brown trout. In: Symposium on DNA-based profiling of mating systems and reproductive behaviors in poikilothermic vertebrates. New Haven, Ct, Jun 17–20 2000, pp 146–149
Gomez-Uchida D, Knight TW, Ruzzante DE (2009) Interaction of landscape and life history attributes on genetic diversity, neutral divergence and gene flow in a pristine community of salmonids. Mol Ecol 18(23):4854–4869. doi:10.1111/j.1365-294X.2009.04409.x
Gomez-Uchida D, Palstra FP, Knight TW, Ruzzante DE (2013) Contemporary effective population and metapopulation size (Ne and meta-Ne): comparison among three salmonids inhabiting a fragmented system and differing in gene flow and its asymmetries. Ecol Evol 3(3):569–580. doi:10.1002/ece3.485
Grenouillet G, Brosse S, Tudesque L, Lek S, Baraillé Y, Loot G (2008) Concordance among stream assemblages and spatial autocorrelation along a fragmented gradient. Divers Distrib 14(4):592–603. doi:10.1111/j.1472-4642.2007.00443.x
Griffiths AM, Ellis JS, Clifton-Dey D, Machado-Schiaffino G, Bright D, Garcia-Vazquez E, Stevens JR (2011) Restoration versus recolonisation: the origin of Atlantic salmon (Salmo salar L.) currently in the River Thames. Biol Conserv 144(11):2733–2738. doi:10.1016/j.biocon.2011.07.017
Hall C, Jordaan A, Frisk M (2011) The historic influence of dams on diadromous fish habitat with a focus on river herring and hydrologic longitudinal connectivity. Landsc Ecol 26(1):95–107. doi:10.1007/s10980-010-9539-1
Hall CJ, Jordaan A, Frisk MG (2012) Centuries of anadromous forage fish loss: consequences for ecosystem connectivity and productivity. Bioscience 62(8):723–731. doi:10.1525/bio.2012.62.8.5
Hansen MM, Nielsen EE, Mensberg KLD (1997) The problem of sampling families rather than populations: relatedness among individuals in samples of juvenile brown trout Salmo trutta L. Mol Ecol 6(5):469–474. doi:10.1046/j.1365-294X.1997.t01-1-00202.x
Hansen MM, Fraser DJ, Meier K, Mensberg KLD (2009) Sixty years of anthropogenic pressure: a spatio-temporal genetic analysis of brown trout populations subject to stocking and population declines. Mol Ecol 18:2549–2562
Haro A, Castro-Santos T, Noreika J, Odeh M (2004) Swimming performance of upstream migrant fishes in open-channel flow: a new approach to predicting passage through velocity barriers. Can J Fish Aquat Sci 61(9):1590–1601
Hendry AP (2004) Selection against migrants contributes to the rapid evolution of ecologically dependent reproductive isolation. Evol Ecol Res 6(8):1219–1236
Horreo JL, Machado-Schiaffino G, Griffiths A, Bright D, Stevens J, Garcia-Vazquez E (2008) Identification of differential broodstock contribution affecting genetic variability in hatchery stocks of Atlantic salmon (Salmo salar). Aquaculture 280(1–4):89–93. doi:10.1016/j.aquaculture.2008.05.004
Horreo JL, Martinez JL, Ayllon F, Pola IG, Monteoliva JA, Heland M, Garcia-Vazquez E (2011) Impact of habitat fragmentation on the genetics of populations in dendritic landscapes. Freshw Biol 56(12):2567–2579. doi:10.1111/j.1365-2427.2011.02682.x
Ikediashi C, Billington S, Stevens JR (2012) The origins of Atlantic salmon (Salmo salar L.) recolonizing the River Mersey in northwest England. Ecol Evol 2(10):2537–2548. doi:10.1002/ece3.353
Johnstone DL, O’Connell MF, Palstra FP, Ruzzante DE (2013) Mature male parr contribution to the effective size of an anadromous Atlantic salmon (Salmo salar) population over 30 years. Mol Ecol 22(9):2394–2407. doi:10.1111/mec.12186
Jones MW, Hutchings JA (2001) The influence of male parr body size and mate competition on fertilization success and effective population size in Atlantic salmon. Heredity (Edinb) 86(Pt 6):675–684
Jonsson B, Jonsson N (2006) Cultured Atlantic salmon in nature: a review of their ecology and interaction with wild fish. In: ICES/NASCO symposium on interactions between aquaculture and wild stocks of atlantic salmon and other diadromous fish species. Bergen, Norway, Oct 18–21 2006, pp 1162–1181. doi:10.1016/j.icesjms.2006.03.004
Jonsson B, Jonsson N (2011) Migrations ecology of Atlantic salmon and brown trout. In: vol 33 fish & fisheries series. Springer, Netherlands, pp 247–325. doi:10.1007/978-94-007-1189-1_6
Kalinowski ST (2005) Hp-rare 1.0: a computer program for performing rarefaction on measures of allelic richness. Mol Ecol Notes 5(1):187–189
Kalinowski ST (2010) The computer program STRUCTURE does not reliably identify the main genetic clusters within species: simulations and implications for human population structure. Heredity 106(4):625–632
Kalinowski ST, Wagner AP, Taper ML (2006) ml-relate: a computer program for maximum likelihood estimation of relatedness and relationship. Mol Ecol Notes 6(2):576–579. doi:10.1111/j.1471-8286.2006.01256.x
Kiffney PM, Pess GR, Anderson JH, Faulds P, Burton K, Riley SC (2009) Changes in fish communities following recolonization of the Cedar River, Wa, USA by pacific salmon after 103 years of local extirpation. River Res Appl 25:438–452
King TL, Eackless MS, Letcher BH (2005) Primer note microsatellite DNA markers for the study of Atlantic salmon (Salmo salar) kinship, population structure, and mixed-fishery analyses. Mol Ecol Notes 5:130–132
Kuparinen A, Tufto J, Consuegra S, Hindar K, Merilä J, de Leaniz CG (2010) Effective size of an Atlantic salmon (Salmo salar L.) metapopulation in Northern Spain. Conserv Genet 11(4):1559–1565
Lange F, Prevost E, Brun M (2011) Les populations de saumons, truites de mer et grandes aloses de la Nivelle en 2010. INRA, St Pée sur Nivelle
Larinier M, Boyer-Bernard S (1991) Dévalaison des smolts et efficacité d’un exutoire de dévalaison à l’usine hydroélectrique d’Halsou sur la Nive. Bull Fr Peche Piscic 321:72–92
Mallen-Cooper M, Brand DA (2007) Non-salmonids in a salmonid fishway: what do 50 years of data tell us about past and future fish passage? Fish Manag Ecol 14(5):319–332. doi:10.1111/j.1365-2400.2007.00557.x
Marie AD, Bernatchez L, Garant D (2010) Loss of genetic integrity correlates with stocking intensity in brook charr (Salvelinus fontinalis). Mol Ecol 19:2025–2037
Martinez JL, Moran P, Perez J, De Gaudemar B, Beall E, Garcia-Vazquez E (2000) Multiple paternity increases effective size of southern Atlantic salmon populations. Mol Ecol 9(3):293–298
Marty A (1984) Le saumon dans les basins de l’Adour et de la Nivelle. Synthesis report ONEMA, Pau
Marty A, Bousquet B (2001) SITUATION DES POISSONS MIGRATEURS AMPHIHALINS SUR LE BASSIN DE L'ADOUR. Bulletin Français de La Peche et de la Piciculture. (357–360):345–356. doi:10.1051/kmae/2001054
McConnell SKJ, O’Reilly P, Hamilton L, Wright JM, Bentzen P (1995) Polymorphic microsatellite loci from Atlantic Salmon (Salmo salar): genetic differentiation of North American and European populations. Can J Fish Aquat Sci 52:1863–1872
McGinnity P, de Eyto E, Cross TF, Coughlan J, Whelan K, Ferguson A (2007) Population specific smolt development, migration and maturity schedules in Atlantic salmon in a natural river environment. Aquaculture 273(2–3):257–268. doi:10.1016/j.aquaculture.2007.10.008
Milot E, Perrier C, Papillon L, Dodson JJ, Bernatchez L (2013) Reduced fitness of Atlantic salmon released in the wild after one generation of captive breeding. Evol Appl 6(3):472–485. doi:10.1111/eva.12028
Moran P, Pendas AM, Beall E, GarciaVazquez E (1996) Genetic assessment of the reproductive success of Atlantic salmon precocious parr by means of VNTR loci. Heredity 77(6):655–660. doi:10.1038/Hdy.1996.193
Naish KA, Seamons TR, Dauer MB, Hauser L, Quinn TP (2013) Relationship between effective population size, inbreeding and adult fitness-related traits in a steelhead (Oncorhynchus mykiss) population released in the wild. Mol Ecol. doi:10.1111/mec.12185
Nei M, Tajima F, Tateno Y (1983) Accuracy of estimated phylogenetic trees from molecular-data.2. Gene-frequency data. J Mol Evol 19(2):153–170
Neraas LP, Spruell P (2001) Fragmentation of riverine systems: the genetic effects of dams on bull trout (Salvelinus confluentus) in the Clark Fork River system. Mol Ecol 10:1153–1164
Nikolic N, Butler JRA, Bagliniere JL, Laughton R, McMyn IAG, Chevalet C (2009) An examination of genetic diversity and effective population size in Atlantic salmon populations. Genet Res 91(6):395–412. doi:10.1017/s0016672309990346
Nilsson C, Reidy CA, Dynesius M, Revenga C (2005) Fragmentation and flow regulation of the world’s large river systems. Science 308(5720):405–408. doi:10.1126/science.1107887
O’Reilly PT, Hamilton L, McConnell SKJ, Wright JM (1996) Rapid analysis of genetic variation in Atlantic salmon (Salmo salar) by PCR multiplexing of dinucleotide and tetranucleotide microsatellites. Can J Fish Aquat Sci 53:2292–2298
Oakley CG (2013) Small effective size limits performance in a novel environment. Evol Appl 6(5):823–831. doi:10.1111/Eva.12068
Oldani NO, Baigún CRM (2002) Performance of a fishway system in a major South American dam on the Parana River (Argentina–Paraguay). River Res Appl 18(2):171–183. doi:10.1002/rra.640
Olson-Manning CF, Wagner MR, Mitchell-Olds T (2012) Adaptive evolution: evaluating empirical support for theoretical predictions. Nat Rev Genet 13(12):867–877. doi:10.1038/nrg3322
Van Oosterhout C, Hutchinson WF, Wills DPM, Shipley P (2004) Micro-checker: software for identifying and correcting genotyping errors in microsatellite data. Mol Ecol Notes 4:535–538
Page RDM (1996) TreeView: an application to display phylogenetic trees on personal computers. Comput Appl Biosci 12(4):357–358
Palstra FP, Ruzzante DE (2011) Demographic and genetic factors shaping contemporary metapopulation effective size and its empirical estimation in salmonid fish. Heredity 107(5):444–455. http://www.nature.com/hdy/journal/v107/n5/suppinfo/hdy201131s1.html
Parrish DL, Behnke RJ, Gephard SR, McCormick SD, Reeves GH (1998) Why aren’t there more Atlantic salmon (Salmo salar)? Can J Fish Aquat Sci 55(S1):281–287. doi:10.1139/d98-012
Paterson S, Piertney SB, Knox D, Gilbey J, Verspoor E (2004) Characterization and PCR multiplexing of novel highly variable tetranucleotide Atlantic salmon (Salmo salar L.) microsatellites. Mol Ecol Notes 4:160–162
Pearse DE, Martinez E, Garza JC (2011) Disruption of historical patterns of isolation by distance in coastal steelhead. Conserv Genet 12(3):691–700. doi:10.1007/s10592-010-0175-8
Pedersen S, Rasmussen G, Nielsen EE, Karlsson L, Nyberg P (2007) Straying of Atlantic salmon, Salmo salar, from delayed and coastal releases in the Baltic Sea, with special focus on the Swedish west coast. Fish Manag Ecol 14(1):21–32
Pépino M, Rodríguez MA, Magnan P (2012) Fish dispersal in fragmented landscapes: a modeling framework for quantifying the permeability of structural barriers. Ecol Appl 22(5):1435–1445. doi:10.1890/11-1866.1
Perrier C, Evanno G, Belliard J, Guyomard R, Bagliniere JL (2010) Natural recolonization of the Seine River by Atlantic salmon (Salmo salar) of multiple origins. Can J Fish Aquat Sci 67(1):1–4. doi:10.1139/F09-190
Perrier C, Grandjean F, Le Gentil J, Cherbonnel C, Evanno G (2011a) A species-specific microsatellite marker to discriminate European Atglantic salmon, brown trout, and their hybrids. Conserv Genet Resour 3(1):131–133
Perrier C, Guyomard R, Bagliniere J-L, Evanno G (2011b) Determinants of hierarchical genetic structure in Atlantic salmon populations: environmental factors vs. anthropogenic influences. Mol Ecol 20(20):4231–4245. doi:10.1111/j.1365-294X.2011.05266.x
Perrier C, Baglinière J-L, Evanno G (2013a) Understanding admixture patterns in supplemented populations: a case study combining molecular analyses and temporally explicit simulations in Atlantic salmon. Evol Appl 6(2):218–230. doi:10.1111/j.1752-4571.2012.00280.x
Perrier C, Guyomard R, Bagliniere J-L, Nikolic N, Evanno G (2013b) Changes in the genetic structure of Atlantic salmon populations over four decades reveal substantial impacts of stocking and potential resiliency. Ecol Evol 3(7):2334–2349. doi:10.1002/ece3.629
Piry S, Alapetite A, Cornuet JM, Paetkau D, Baudouin L, Estoup A (2004) GENECLASS2: a software for genetic assignment and first-generation migrant detection. J Hered 95(6):536–539. doi:10.1093/jhered/esh074
Poulet N (2007) Impact of weirs on fish communities in a piedmont stream. River Res Appl 23(9):1038–1047. doi:10.1002/rra.1040
Primmer CR (2011) Genetics of local adaptation in salmonid fishes. Heredity 106(3):401–403
Primmer CR, Veselov AJ, Zubchenko A, Poututkin A, Bakhmet I, Koskinen MT (2006) Isolation by distance within a river system: genetic population structuring of Atlantic salmon, Salmo salar, in tributaries of the Varzuga River in northwest Russia. Mol Ecol 15:653–666
Pritchard JK, Stephens P, Donelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Prouzet P (1990) Stock characteristics of Atlantic salmon Salmo salar in france a review. Aquat Living Resour 3(2):85–98
Quinn TP (1993) a review of homing and straying of wild and hatchery-produced salmon. Fish Res 18:29–44
Raeymaekers JAM, Raeymaekers D, Koizumi I, Geldof S, Volckaert FAM (2009) Guidelines for restoring connectivity around water mills: a population genetic approach to the management of riverine fish. J Appl Ecol 46(3):562–571. doi:10.1111/j.1365-2664.2009.01652.x
Raymond M, Rousset F (1995a) An exact test for population differentiation. Evolution 49:1280–1283
Raymond M, Rousset F (1995b) GENEPOP (version 1.2): population genetics software for exact tests and ecumenicism. J Hered 86:248–249
Richard A, Dionne M, Wang J, Bernatchez L (2013) Does catch and release affect the mating system and individual reproductive success of wild Atlantic salmon (Salmo salar L.)? Mol Ecol 22(1):187–200. doi:10.1111/mec.12102
Saura M, Caballero A, Caballero P, Moran P (2008) Impact of precocious male parr on the effective size of a wild population of Atlantic salmon. Freshw Biol 53(12):2375–2384. doi:10.1111/j.1365-2427.2008.02062.x
Schreiber A, Diefenbach G (2005) Population genetics of the European trout (Salmo trutta L.) migration system in the river Rhine: recolonisation by sea trout. Ecol Freshw Fish 14(1):1–13. doi:10.1111/j.1600-0633.2004.00072.x
Slettan A, Olsaker I, Lie O (1995) Atlantic Salmon microsatellites at the SSOSL25, SSOSL85 SSOSL311 and SSOSL417 loci. Anim Genet 26:281
Stabell OB (1984) Homing and olfaction in Salmonids—a critical review with special reference to the Atlantic salmon. Biol Rev Camb Philos Soc 59:333–388
Taylor EB (1991) A review of local adaptation in salmonidae, with particular reference to pacific and Atlantic salmon. Aquaculture 98(1–3):185–207
Thériault V, Moyer GR, Jackson LS, Blouin MS, Banks MA (2011) Reduced reproductive success of hatchery coho salmon in the wild: insights into most likely mechanisms. Mol Ecol 20(9):1860–1869. doi:10.1111/j.1365-294X.2011.05058.x
Tonteri A, Veselov AJ, Zubchenko AV, Lumme J, Primmer CR (2009) Microsatellites reveal clear genetic boundaries among Atlantic salmon (Salmo salar) populations from the Barents and White seas, northwest Russia. Can J Fish Aquat Sci 66(5):717–735. doi:10.1139/f09-010
Traill LW, Brook BW, Frankham RR, Bradshaw CJA (2010) Pragmatic population viability targets in a rapidly changing world. Biol Conserv 143(1):28–34. doi:10.1016/j.biocon.2009.09.001
Tsuboi J-I, Iwata T, Morita K, Endou S, Oohama H, Kaji K (2013) Strategies for the conservation and management of isolated salmonid populations: lessons from Japanese streams. Freshw Biol 58(5):908–917. doi:10.1111/fwb.12096
Vaha JP, Erkinaro J, Niemela E, Primmer CR (2007) Life-history and habitat features influence the within-river genetic structure of Atlantic salmon. Mol Ecol 16(13):2638–2654. doi:10.1111/j.1365-294X.2007.03329.x
Vaha JP, Erkinaro J, Niemela E, Primmer CR (2008) Temporally stable genetic structure and low migration in an Atlantic salmon population complex: implications for conservation and management. Evol Appl 1(1):137–154. doi:10.1111/j.1752-4571.2007.00007.x
Van Oort H, McLellan BN, Serrouya R (2011) Fragmentation, dispersal and metapopulation function in remnant populations of endangered mountain caribou. Anim Conserv 14(3):215–224. doi:10.1111/j.1469-1795.2010.00423.x
Van Oosterhout C, Hutchinson WF, Wills DPM, Shipley P (2004) MICRO-CHECKER: software for identifying and correcting genotyping errors in microsatellite data. Mol Ecol Notes 4(3):535–538. doi:10.1111/j.1471-8286.2004.00684.x
Vauclin V (2007) French Implementation plan of NASCO’s resolutions and agreements with regard to the protection, the management and the exploitation of the Atlantic salmon and its Habitats. Report ONEMA, Orléans
Vitousek PM, Mooney HA, Lubchenco J, Melillo JM (1997) Human domination of earth’s ecosystems. Science 277(5325):494–499. doi:10.1126/science.277.5325.494
Waples RS, Do C (2008) ldne: a program for estimating effective population size from data on linkage disequilibrium. Mol Ecol Resour 8(4):753–756. doi:10.1111/j.1755-0998.2007.02061.x
Waples RS, England PR (2011) Estimating contemporary effective population size on the basis of linkage disequilibrium in the face of migration. Genetics 189(2):633–644. doi:10.1534/genetics.111.132233
Whiteley AR, Coombs JA, Hudy M, Robinson Z, Colton AR, Nislow KH, Letcher BH (2013) Fragmentation and patch size shape genetic structure of brook trout populations. Can J Fish Aquat Sci 70(5):678–688. doi:10.1139/cjfas-2012-0493
Winans GA, Baird MC, Baker J (2010) A genetic and phenetic baseline before the recolonization of steelhead above Howard Hanson Dam, Green River, Washington. North Am J Fish Manag 30(3):742–756
Wofford JEB, Gresswell RE, Banks MA (2005) Influence of barriers to movement on within-watershed genetic variation of coastal cutthroat trout. Ecol Appl 15(2):628–637. doi:10.1890/04-0095
Acknowledgments
We acknowledge all participants to the collection of samples and of various historical and environmental data, with a special attention to A. Manicki, J. Chat, D. Barracou. We also thank P. Regnacq, JB Torterotot and Anne Dalziel for their help while analyzing data and writing the paper. We thank two anonymous reviewers and the associate editor C. Primmer for their very constructive comments. Authors also thank all French organizations that provided their technical assistance for electric fishing: the National Institute for Agricultural Research (INRA), the National Office of Water and Aquatic Media (ONEMA) and Migradour. This work was funded by the European Union INTERREG IIIB program [Atlantic Salmon Arc Project (ASAP)] and the European Union INTERREG IVB program [Atlantic Arc Resource Conservation (AARC)].
Author information
Authors and Affiliations
Corresponding author
Additional information
Charles Perrier and Jérôme Le Gentil have contributed equally to this study.
Rights and permissions
About this article
Cite this article
Perrier, C., Le Gentil, J., Ravigne, V. et al. Origins and genetic diversity among Atlantic salmon recolonizing upstream areas of a large South European river following restoration of connectivity and stocking. Conserv Genet 15, 1095–1109 (2014). https://doi.org/10.1007/s10592-014-0602-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10592-014-0602-3