Abstract
Purpose
The purpose of this study is to characterize a novel structural variant, a large duplication involving exons 1–19 of the BRCA1 gene in four independent families, and to provide diagnostically valuable information including the position of the breakpoints as well as clues to its clinical significance.
Methods
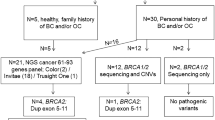
The duplication of exons 1–19 of the BRCA1 gene was initially detected by routine laboratory testing including MLPA analysis and next generation sequencing. For detailed characterization we performed array-comparative genome hybridization analysis, fluorescent in situ hybridization, next generation mapping, and long-distance PCR for break-point sequencing.
Results
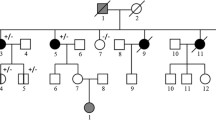
Our data revealed a tandem duplication on chromosome 17 that encompassed 357 kb and included exons 1–19 of the BRCA1 gene and the genes NBR2, NBR1, TMEM106A, LOC100130581, ARL4D, MIR2117 up to parts of the DHX8 gene. This structural variant appeared as a tandem duplication with breakpoints in intron 19 of the BRCA1 gene and in intron 3 of the DHX8 gene (HGVS:chr17(hg19):g.41210776_41568516dup). Segregation analysis indicated that this structural rearrangement is phased in trans with a known pathogenic exon deletion of the BRCA1 gene in one family.
Conclusions
The copy number variation initially recognized as duplication of exon 1–19 of the BRCA1 gene by MLPA analysis is a structural variation with breakpoints in the BRCA1 and DHX8 genes. Although currently to be classified as a variant of unknown significance, our family data indicates that this duplication may be a benign variation or at least of markedly reduced penetrance since it occurs in trans with another known fully pathogenic variant in the BRCA1 gene.





Similar content being viewed by others
References
Kuchenbaecker KB, Hopper JL, Barnes DR, Phillips KA, Mooij TM, Roos-Blom MJ, Jervis S, van Leeuwen FE, Milne RL, Andrieu N, Goldgar DE, Terry MB, Rookus MA, Easton DF, Antoniou AC, Brca, Consortium BC, McGuffog L, Evans DG, Barrowdale D, Frost D, Adlard J, Ong KR, Izatt L, Tischkowitz M, Eeles R, Davidson R, Hodgson S, Ellis S, Nogues C, Lasset C, Stoppa-Lyonnet D, Fricker JP, Faivre L, Berthet P, Hooning MJ, van der Kolk LE, Kets CM, Adank MA, John EM, Chung WK, Andrulis IL, Southey M, Daly MB, Buys SS, Osorio A, Engel C, Kast K, Schmutzler RK, Caldes T, Jakubowska A, Simard J, Friedlander ML, McLachlan SA, Machackova E, Foretova L, Tan YY, Singer CF, Olah E, Gerdes AM, Arver B, Olsson H (2017) Risks of breast, ovarian, and contralateral breast cancer for BRCA1 and BRCA2 mutation carriers. JAMA 317(23):2402–2416. https://doi.org/10.1001/jama.2017.7112
Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, Grody WW, Hegde M, Lyon E, Spector E, Voelkerding K, Rehm HL, Committee ALQA (2015) Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med 17(5):405–424. https://doi.org/10.1038/gim.2015.30
Ready K, Gutierrez-Barrera AM, Amos C, Meric-Bernstam F, Lu K, Hortobagyi G, Arun B (2011) Cancer risk management decisions of women with BRCA1 or BRCA2 variants of uncertain significance. Breast J 17(2):210–212. https://doi.org/10.1111/j.1524-4741.2010.01055.x
Cheon JY, Mozersky J, Cook-Deegan R (2014) Variants of uncertain significance in BRCA: a harbinger of ethical and policy issues to come? Genome Med 6(12):121. https://doi.org/10.1186/s13073-014-0121-3
Hauke J, Horvath J, Gross E, Gehrig A, Honisch E, Hackmann K, Schmidt G, Arnold N, Faust U, Sutter C, Hentschel J, Wang-Gohrke S, Smogavec M, Weber BHF, Weber-Lassalle N, Weber-Lassalle K, Borde J, Ernst C, Altmuller J, Volk AE, Thiele H, Hubbel V, Nurnberg P, Keupp K, Versmold B, Pohl E, Kubisch C, Grill S, Paul V, Herold N, Lichey N, Rhiem K, Ditsch N, Ruckert C, Wappenschmidt B, Auber B, Rump A, Niederacher D, Haaf T, Ramser J, Dworniczak B, Engel C, Meindl A, Schmutzler RK, Hahnen E (2018) Gene panel testing of 5589 BRCA1/2-negative index patients with breast cancer in a routine diagnostic setting: results of the German Consortium for Hereditary Breast and Ovarian Cancer. Cancer Med. https://doi.org/10.1002/cam4.1376
Gadzicki D, Evans DG, Harris H, Julian-Reynier C, Nippert I, Schmidtke J, Tibben A, van Asperen CJ, Schlegelberger B (2011) Genetic testing for familial/hereditary breast cancer-comparison of guidelines and recommendations from the UK, France, the Netherlands and Germany. J Community Genet 2:53–69
Petrucelli N, Daly MB, Pal T (2016) BRCA1- and BRCA2-Associated Hereditary Breast and Ovarian Cancer. In: Adam MP, Ardinger HH, Pagon RA et al (eds) GeneReviews® [Internet]. University of Washington, Seattle(WA). Available from: https://www.ncbi.nlm.nih.gov/books/NBK1247/
Domchek SM, Tang J, Stopfer J, Lilli DR, Hamel N, Tischkowitz M, Monteiro AN, Messick TE, Powers J, Yonker A, Couch FJ, Goldgar DE, Davidson HR, Nathanson KL, Foulkes WD, Greenberg RA (2013) Biallelic deleterious BRCA1 mutations in a woman with early-onset ovarian cancer. Cancer Discov 3(4):399–405. https://doi.org/10.1158/2159-8290.CD-12-0421
Sawyer SL, Tian L, Kahkonen M, Schwartzentruber J, Kircher M, University of Washington Centre for Mendelian G, Consortium FC, Majewski J, Dyment DA, Innes AM, Boycott KM, Moreau LA, Moilanen JS, Greenberg RA (2015) Biallelic mutations in BRCA1 cause a new Fanconi anemia subtype. Cancer Discov 5(2):135–142. https://doi.org/10.1158/2159-8290.CD-14-1156
Wong-Brown M, McPhillips M, Gleeson M, Spigelman AD, Meldrum CJ, Dooley S, Scott RJ (2016) When is a mutation not a mutation: the case of the c.594-2A> C splice variant in a woman harbouring another BRCA1 mutation in trans. Hered Cancer Clin Pract 14:6. https://doi.org/10.1186/s13053-015-0045-y
Janavicius R (2010) Founder BRCA1/2 mutations in the Europe: implications for hereditary breast-ovarian cancer prevention and control. EPMA J 1(3):397–412. https://doi.org/10.1007/s13167-010-0037-y
Liu X, Xiao ZD, Han L, Zhang J, Lee SW, Wang W, Lee H, Zhuang L, Chen J, Lin HK, Wang J, Liang H, Gan B (2016) LncRNA NBR2 engages a metabolic checkpoint by regulating AMPK under energy stress. Nat Cell Biol 18(4):431–442. https://doi.org/10.1038/ncb3328
Deosaran E, Larsen KB, Hua R, Sargent G, Wang Y, Kim S, Lamark T, Jauregui M, Law K, Lippincott-Schwartz J, Brech A, Johansen T, Kim PK (2013) NBR1 acts as an autophagy receptor for peroxisomes. J Cell Sci 126(Pt 4):939–952. https://doi.org/10.1242/jcs.114819
Smith SA, Holik PR, Stevens J, Melis R, White R, Albertsen H (1995) Isolation and mapping of a gene encoding a novel human ADP-ribosylation factor on chromosome 17q12-q21. Genomics 28(1):113–115. https://doi.org/10.1006/geno.1995.1115
Ohno M, Shimura Y (1996) A human RNA helicase-like protein, HRH1, facilitates nuclear export of spliced mRNA by releasing the RNA from the spliceosome. Genes Dev 10(8):997–1007
Chiang AP, Nishimura D, Searby C, Elbedour K, Carmi R, Ferguson AL, Secrist J, Braun T, Casavant T, Stone EM, Sheffield VC (2004) Comparative genomic analysis identifies an ADP-ribosylation factor-like gene as the cause of Bardet-Biedl syndrome (BBS3). Am J Hum Genet 75(3):475–484. https://doi.org/10.1086/423903
Acknowledgements
The authors would like to thank the patients and their families for supporting this study and Marcel Tauscher and Hildegard Frye-Boukhriss for excellent technical assistance.
Funding
This work was funded in parts by the German Research Foundation (DFG), Cluster of Excellence REBIRTH to B.S. and D.S.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Disclosures
SC and HC hold stocks of Bionano Genomics, Inc. All other authors have no conflict of interest to declare.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Du, C., Mark, D., Wappenschmidt, B. et al. A tandem duplication of BRCA1 exons 1–19 through DHX8 exon 2 in four families with hereditary breast and ovarian cancer syndrome. Breast Cancer Res Treat 172, 561–569 (2018). https://doi.org/10.1007/s10549-018-4957-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10549-018-4957-x