Abstract
Conservation of imperiled plant species often requires ex situ (offsite) living collections. Protocols for developing these collections most often emphasize sampling depth, but little is known about the genetics of such collections. This study compares how well a single collecting protocol can capture the diversity in wild populations of two closely related species. We selected two exemplar species, bay rush (Zamia lucayana) and sinkhole cycad (Zamia decumbens), based on similarities and differences that allow for rigorous comparison, including geographic isolation and reproductive factors. For each species, we compared in situ plants to ex situ plants via the same panel of 10 microsatellite markers. Genetic distance analysis shows high fidelity of the ex situ collections to their in situ source populations and sub-populations. Structured resampling of allele capture from the in situ populations by the ex situ collections shows that allele capture increases as number of ex situ plants maintained increases, but with a diminishing rate of increase. Difference in the rate of allele capture between the two species was significant at the α = 0.1 level, (p = 0.097) but not at the α = 0.05 level. At larger collection sizes, the difference in rate of allele capture showed a high practical significance (d = 5.41). These data illustrate that a unified collecting protocol can achieve similar allele capture among related species, but also that geographic and reproductive factors can influence the rate of allele capture.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Ex situ botanic garden conservation
Botanic gardens increasingly emphasize conservation in their work (Havens et al. 2014). One of the most central contributions botanic gardens make towards conservation is the offsite protection of imperiled plant species (Larkin et al. 2016; Smith et al. 2016). In addition to seed banking and tissue culture (Pence 2013; Berjak and Pammenter 2014; Raven and Havens 2014), cultivation of living collections is an effective method for preserving threatened plant species (Gemmill et al. 1998; Cibrian-Jaramillo et al. 2013; Cavender et al. 2015). Ex situ conservation can be central to species recovery efforts (Arnet et al. 2015). Including ex situ collections in conservation planning can increase the probability of successful recovery (McGowen et al. 2016).
Sampling strategies for genetic capture
Much progress has been made in understanding the sampling strategies for living conservation collections, mostly for seed storage (Schoen and Brown 2001; Guerrant et al. 2004, 2014; McGlaughlin et al. 2015) and also with regard to sampling breadth (Larkin et al. 2016). This new understanding for botanic garden work has a basis in the agricultural sciences (Frankel 1984; Gale and Lawrence 1984; Brown 1989) which could be more vigorously applied (Heywood 2009). Recent investigation into the genetic basis of sampling strategies offers finer insight on how effective such strategies can be (Richards et al. 2007; Brütting et al. 2013). Sufficient sampling depth within a population is an essential starting point, but careful attention to biological and geographic factors can lead to more effective protocols (Hoban and Schlarbaum 2014; Hoban and Strand 2015).
Ex situ collections may differ in effectiveness
Living ex situ collections are central to conservation planning for certain plants, including cycads (Walters 2003; Okubamichael et al. 2016). Recent population genetic study suggests that a much greater sampling depth is required to capture sufficient genetic diversity of cycads than the depth required for other plant groups (BGCI 2014; Griffith et al. 2015). While these types of empirical assays offer much needed confirmation and refinement of existing protocols, an admitted limit is the unknown breadth of applicability of such assays, even among closely related species (Namoff et al. 2010; Griffith and Husby 2010).
Thus, the current study seeks to compare how broadly such findings apply. Stated as a question: Will a single conservation sampling protocol, such as that put forward in Griffith et al. (2015), capture similar genetic diversity between two related species? Or will a single protocol capture genetic diversity at different levels based on biological factors? Generation time and reproductive frequency, for example, can influence genetic structure (Hamrick and Godt 1996; Duminil et al. 2007), and thus may influence how well a collection protocol may capture in situ diversity. We hypothesize that when using the same collection protocol, the rate of genetic capture as a function of sample size will differ, even among closely related species. A comparison of the population genetics of ex situ collections of two related species can explore this hypothesis. To our knowledge, this is the first such study to compare two closely-related real-world examples, carefully selected to enable comparisons of geographic and biological factors, and utilizing extensive sampling of both in situ and ex situ genetic diversity.
Materials and methods
Model system
For this study, we examine the effectiveness of ex situ conservation protocols at Montgomery Botanical Center (MBC; Coral Gables, Florida, USA) in capturing in situ genetic diversity. MBC cycad collections are curated to maximize genetic diversity at the population level (Calonje et al. 2009a), balanced with resource, space and logistic limitations (Griffith and Husby 2010). This curation strategy is based on the well-understood negative influence on conservation from inbreeding depression (Schemske et al. 1994; Frankham 1995) or loss of diversity through drift (Gale and Lawrence 1984). Estimates of genetic diversity from allozyme data (Walters and Decker-Walters 1991) informed a long-standing population-based collecting protocol at MBC, which sought to maintain at least 15 plants from each population, grown from wild-collected seed from at least 3 mother plants. Based on recent microsatellite assay (Griffith et al. 2015), current cycad collecting protocols now seek to maintain as many (seed-grown) plants from as broad a group of mother plants as possible, without negatively impacting the source population (Menges et al. 2004).
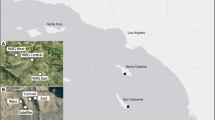
The current study explores genetic diversity of bay rush (Zamia lucayana Britton: Zamiaceae). Zamia lucayana is endemic to a single Caribbean island (Long Island, The Bahamas), where it is restricted to a narrow strip of coastal sand dunes (Fig. 1). This species is now considered critically endangered due to small population size, limited extent of occurrence, and pressure from residential development and sand mining (Calonje et al. 2013). Conservation genetic analysis indicates that Z. lucayana has significant genetic structure as seen via analysis of molecular variance, but is considered a single population for management purposes (Calonje et al. 2013). Zamia species, and cycads in general are ideal candidates for ex situ conservation given slow reproductive cycles, concerns with poaching, and recalcitrant seeds (BGCI 2015).
Model system used in this study. Bay rush (a Zamia lucayana) is restricted to a single continuous population on Long Island, The Bahamas. The two major sinkhole cycad (Zamia decumbens) populations are restricted to two large limestone sinkholes in southern Belize (b). These in situ plants (a, b) were compared to ex situ plants at Montgomery Botanical Center (c), which were grown from seed collected in 2009 (bay rush, upper right), and 2010 (sinkhole cycad, upper left)
We selected bay rush for comparison with sinkhole cycad (Zamia decumbens Calonje, Meerman, M. P.Griff. and Hoese: Zamiaceae) investigated in Griffith et al. 2015) because of similarities and differences between the two species (Fig. 1). The important similarities are their close congeneric status and their shared critically endangered status (Calonje et al. 2016). The following specific circumstances set bay rush distinctly apart from sinkhole cycad. Geography: bay rush has an extremely small range, with a single population restricted to a short, narrow band of coastal sand dunes, whereas sinkhole cycad has a much broader distribution, occurring in two disjunct populations in Southern Belize. Population size: bay rush has a single population of ca. 1000 plants (Calonje et al. 2013), whereas sinkhole cycad populations do not exceed 200 plants, and less than 500 plants are known (Calonje et al. 2009b). Reproductive differences: Caribbean zamias (from Florida and the West Indies), such as bay rush, have distinctively different life histories than rainforest cycads such as sinkhole cycad, as Caribbean zamias reach reproductive maturity faster and produce reproductive structures more often than rainforest zamias (Griffith et al. 2012; Clugston et al. in press). Environmental differences: bay rush occurs in full sun on coastal white limestone sand dunes, whereas sinkhole cycad occurs in deep shade at the bottom of humus-rich sinkholes. These environmental differences can influence gene flow, fecundity, germination, seedling recruitment, and survival (Clark and Clark 1987; Lopez-Gallego and O’Neil 2010) and therefore alter the demographic structure of populations. In addition to these biological and geographic factors, we selected these two species for logistic reasons. Both are especially well suited for the current study due to abundant, well curated ex situ collections available, and detailed documentation of provenance (Calonje et al. 2009b, 2013).
Sampling protocol
Thorough sampling of leaflets of in situ individuals from throughout the native populations was performed, as detailed in Calonje et al. (2013) and Griffith et al. (2015), and summarized here and in Table 1. Leaflet samples from dispersed individuals covering the geographic span of the subpopulations were collected and labelled, with an emphasis on even spatial distribution. This study compares the in situ bay rush samples to cultivated plants in MBC ex situ collections derived from the in situ population (Table 1). The ex situ collection is seed-grown from in situ female plants. Within the single population, bay rush has three major sub-populations, termed Buckley’s, Hamilton’s and Petty’s (Calonje et al. 2013). We compared these three in situ sub-populations to living collections developed from seeds collected during fieldwork in 2009. The ex situ plants are curated as separate accessions, defined as collections derived from single, separate mother plants (7 accessions from Buckley’s, 5 accessions from Hamilton’s, and 4 accessions from Petty’s; see Table 1).
DNA extraction and amplification
DNA isolation, PCR amplification and visualization of SSR fragments follow protocols described by Meerow and Nakamura (2007). We used ten microsatellite primers for this analysis: Zam28, Zam33, Zam53, Zam59, Zam60, Zam61, Zfg23, Zfg25, Zfg32, and Zfg33 (Meerow et al. 2012). These neutral molecular markers are the same microsatellites used in Griffith et al. (2015), allowing for precise comparison between bay rush and sinkhole cycad. The datasets generated and analysed during the current study are available as a supplementary file with the online version of the article.
Population genetic assay and structured resampling
We implemented comparative estimates of genetic distance (Nei 1978) and multivariate analysis of genetic distance (Orloci 1978; Huff et al. 1993) in GenAlEx version 6.41 (Peakall and Smouse 2006). To determine the level of genetic diversity captured via the population-based collecting protocol, we compared the amount of alleles in the in situ sub-populations and the ex situ collections, based on protocols developed by Namoff et al. (2010). The number of bay rush ex situ samples (n = 244 for all three sub-populations together; Table 1) allows exploration of optimal collection sizes. We resampled the collection data, without replacement, to obtain randomly selected model populations (hereafter referred to as ‘resamples’). These resamples were composed of randomly selected entire accessions, in other words plants derived from one mother, i.e. half-sibling cohorts. For example, one random resample of 51 plants was composed of 19 plants from one mother (accession 14; Table 1), 18 plants from a second mother (accession 16), and 14 plants from a third mother (accession 20), and included all the offspring from these three mother plants, and no offspring from any other mother plant. The resamples were structured to include from one to 16 accessions and from 8 to 244 individuals (i.e. spanning the entire range of the current ex situ collection). We made measures of genetic capture for these random samples by comparing each resample to the population via GenAlEx, and comparing the proportion of private alleles to total alleles in the population and resample. We modeled allelic capture as a function of number of individuals in the collection via a logarithmic regression fit to the allele capture for each species using GenAlEx, giving a curve of expected genetic capture as a function of collection size.
Comparison between species
We designed sampling protocols, collections management, and population genetic assay methods for the current study to mirror the methods used in the prior recent study of the sinkhole cycad (Griffith et al. 2015), allowing for comparison between two congeneric, critically endangered species. We compared allele capture data for significance and effect size (Cohen 1988) using Real Statistics Resource Pack version 4.3 (Zaiontz 2015).
Results
Distance analysis
Genetic distance analysis shows a high degree of identity for each bay rush ex situ collection compared to its source sub-population. The ex situ collections from Hamilton’s, Buckley’s, and Petty’s, in that order, show nearest distance with their respective source sub-populations (Table 2). Multivariate analysis of genetic distance by individual plants also shows high identity of the ex situ collections to their respective source populations, but indicates no clear separation by genetic distance between the sub-populations, (Fig. 2).
Multivariate analysis of genetic distance data for all bay rush (Zamia lucayana) plants in the study (n = 365, see Table 1). The first two axes (PCA 1 and PCA 2) are indicated with the percent of variation explained by each. Each point is an individual plant. Squares indicate in situ (wild) plants (Calonje et al. 2013), and circles represent ex situ plants. Clear overlap among the three in situ sub-populations is noted, yet ex situ collections show high fidelity with the in situ sub-population from which they were collected (Table 2)
Genetic capture by ex situ collection
Including all available accessions, the ex situ collection captures 89.91% of the alleles from the in situ population of bay rush. With each sub-population treated separately, collections from Buckley’s capture 85.18% of in situ alleles, collections from Hamilton’s capture 68.96% of alleles, and collections from Petty’s capture 88.75% of alleles.
Structured resampling of allele capture by collection size shows an increase in genetic capture as collection size increases (Fig. 3). Collections composed of a single accession (i.e. a half-sibling cohort from a single mother plant; 16 possible iterations, 8–28 individual plants) captured between 23.53 and 51.26% of the alleles in the in situ population, while the full ex situ collection of 16 accessions (1 possible iteration, 244 plants) captured 89.91% of the in situ alleles. A proportional decrease in the rate of increase is also indicated through a “diminishing returns” relationship (Fig. 3, r2 = 0.9444), estimated as genetic capture = 0.1973 [ln(collection size)] − 0.1742.
Ex situ collection modeling for bay rush (Zamia lucayana) compared to sinkhole cycad (Zamia decumbens; Griffith et al. 2015), showing % of alleles captured in random resamples of the ex situ collection, by total number of individual plants in the collection. Larger point sizes correspond with larger number of accessions (=more half-sibling cohorts). For bay rush, a single accession collection (smallest black points) captures between 24 and 51% of in situ alleles, while the entire ex situ collection (16 accessions, 244 plants) captures 90% of in situ alleles. The allele capture data are statistically different (p = 0.097; α = 0.1) for the two species, with a large effect size (d = 5.41)
Comparison between species
Comparison of the allele capture data for bay rush with that for sinkhole cycad shows a different (p = 0.0976) rate of genetic capture as a function of collection size (Fig. 3). For a given number of plants, ex situ collections of bay rush capture more in situ allele diversity than collections of sinkhole cycad. A bay rush collection comprised of 3 accessions with 15 mother plants (cf. Namoff et al. 2010) would potentially capture an average of 36.0% of in situ alleles, while a similar collection of sinkhole cycad would capture only an average of 24.3% of the in situ diversity. For comparative purposes, expressed as an 80% target for genetic capture, a sinkhole cycad collection would require at least 190 plants, while a bay rush collection can reach this target with 140 plants. Between these two collection sizes (x̄ = 165 plants) bay rush captures an average of 10.31% more alleles that sinkhole cycad (d = 5.41; 95% confidence interval 3.15, 6.33).
Discussion
Exploring the hypothesis
The title of this paper asks a question which frames this discussion: can ex situ collections conserve different plant species equally well? An intuitive answer might be “no,” given the wide phyletic distances within the plant kingdom, potential great diversity in biology and geography of individual populations (Hoban and Schlarbaum 2014), life histories, threats, and circumstances among plants (Griffith et al. 2011), and the particular challenges in cultivating many species (Calonje et al. 2010). This study seeks a more data-driven answer to that question by isolating many of those variables, and carefully considering the differences that remain between these two empirical cases.
We hypothesize that for the same collection protocol, the rate of genetic capture can differ even between closely related species. For the two species examined here, the null hypothesis (no difference in rate of genetic capture) is not rejected at the 95% confidence level, but is rejected at the 90% confidence level (Fig. 3). Furthermore, the effect size value (d = 5.41) shows a high practical significance at larger collection sizes. Thus, for this discussion, it is useful to consider how these two species behave similarly in ex situ collections, and also how they differ.
Similarities
Comparing the two Zamia species shows that both exhibit a relationship of diminishing returns on genetic capture as a function of collection size, i.e. that the rate of increase in genetic capture decreases as the number of plants increases (Fig. 3). Both species show greatest variation in allele capture for collections composed of a single accession (i.e. half-sibling cohort). As the number of accessions increases, the % allele capture increases and the range of % allele capture decreases. This relationship of diminishing returns has been found in similar studies for unrelated plant species. Richards et al. (2007), also using microsatellite data and similar simulated resamples of a collection of Texas wild rice (Zizania texana), observed the same diminishing returns on allele capture as collection size increased. Volk et al. (2005) show a similar pattern for wild apple (Malus sieversii), also using microsatellite data. A study of Keys thatch palm (Leucothrinax morrisii) also using structured resampling found the same general relationship, (Namoff et al. 2010). The similarities in allele capture between the Zamia species in the current study, and among other species investigated in a similar manner, further validate the emphasis of current collecting protocols to sample for depth within a given population of any species (Guerrant et al. 2004, 2014; Haidet and Olwell, 2015).
Differences
The main difference is that for a given collection size, bay rush collections capture a greater amount of genetic diversity than collections of the sinkhole cycad. Conversely, to achieve a targeted level of allele capture, a greater number of sinkhole cycad collections than bay rush collections would be required, e.g., for 80% allele capture, 190 or 140 plants respectively (Fig. 3). For comparison, 125 plants of Texas wild rice achieved above 80% allele capture (Richards et al. 2007), and a core collection of 15-20 plants of wild apple captures 80% of allele diversity (Volk et al. 2005). In the Keys thatch palm example (using dominant ISSR markers), a collection of only 15 ex situ plants were required to reach 80% allele capture (Namoff et al. 2010). Thus, the idea that every species is different with regard to allele capture in ex situ collections (Griffith et al. 2015) is supported by these data, even between the two closely related Zamia species studied here. By using an identical collecting protocol and genetic assay method, this leaves biological differences as the most likely cause of differences in genetic capture, but sampling error could also be considered (Hong and Ellis 1996). Either way, given the level of resource input required for ex situ collections (Pardey et al. 1998; Cibrian-Jaramillo et al. 2013), these differences have implications for feasibility and management. Thus, these data can inform planning for ex situ conservation collections, if the biological similarities and differences between cases are carefully considered.
Insights from this model system
By examining two closely related species which differ in geographic and reproductive factors, this study provides a comparison which can offer insight for ex situ collections protocols. In this way, we address some limitations expressed in Namoff et al. (2010), which noted that a single species assay offers information, but it is not known how broadly such findings can be applied. By comparison, insights and recommendations can be further refined.
Differences in geographic structure are known to affect genetic structure (Gapare and Aitken 2005; Lopez-Gallego and O’Neil 2010). Thus, geographic structure is considered to affect allele capture for ex situ collections (Touchell et al. 1997; Hoban and Schlarbaum 2014; Hoban and Strand 2015). The case study presented here can empirically illustrate the need to consider geographic structure in collecting protocols by comparing two cycad species with different geographic structure. Bay rush exists as a single continuous population within 7 km, whereas sinkhole cycad is separated into two disjunct major populations 7 km apart. This geographic structure is mirrored in the multivariate analysis of genetic distance data for bay rush, which shows little separation by either axis (Fig. 2), while the same analysis for sinkhole cycad completely separates its two populations via a single axis (Griffith et al. 2015). For bay rush, a lack of clear differentiation by genetic distance justifies treating the in situ plants as a single population for management purposes (Calonje et al. 2013).
Can these differences in genetic and geographic structure inform collecting protocols? This can be answered by separating out the allele capture for selected cohorts within the bay rush ex situ collection, and comparing to sinkhole cycad (Table 3). These data show that collecting from only a single sub-population of bay rush can result in either close to expected or far less than expected genetic capture than from similarly-sized collection of plants from multiple sub-populations. Ex situ collections from Hamilton’s (n = 77), for example, can only capture up to 52% of the genetic diversity of the full in situ population of bay rush, far less than the 68% expected for a collection of 77 plants randomly chosen from all three subpopulations (Table 3). However, ex situ collections from either Buckley’s or Petty’s perform much nearer to expectations (Table 3). A similar, but even more variable result is seen for sinkhole cycad (Table 3); ex situ plants from a single population either capture much fewer (Sinkhole 1), or close to the expected amount of alleles. Thus, the current study exemplifies the need for consideration of geographic factors when developing ex situ collections, and lends support to the idea that every population should be considered separately for such work (Ceska et al. 1997; Krishnan et al. 2013; BGCI 2014). Thus, we recommend carefully considering geographic differences among populations when implementing ex situ conservation actions.
Bay rush and sinkhole cycad also differ in reproductive phenology and life history. As in all cycads, both species are dioecious and pollinated by specialist insects (Rhopalotria dimidiata and R. calonjei, respectively; O’Brien and Tang 2015). Bay rush is observed to have numerous in situ female plants showing multiple cones with complete seed set (Calonje et al. 2013), whereas in a recent observation sinkhole cycad shows much less reproductive frequency, with only 7 plants out of 375 bearing mature seed, and most of these bearing single cones (Griffith et al. 2015). These differences persist in a common garden setting. Bay rush collections begin to produce male cones in 2 years from seed, and female cones in 3 years; sinkhole cycad collections have only produced one male cone in collections 6 years from seed, and no cones in another cohort of collections 8 years from seed. Based on this limited information, the sinkhole cycad has a minimum absolute generation time at least three times as long as bay rush, and wild populations of sinkhole cycad exhibit considerably lower gene flow each year. This correlates with a greater number of seed collections needed to capture sufficient allele diversity for sinkhole cycad (Fig. 3).
Schoen and Brown (1991) as well as Hoban and Strand (2015) simulated the effects of selfing and limited dispersal on seed collection and also found that much greater sampling is needed when these reproductive factors reduce gene flow. While dioecious Zamia are obligate outcrossers, very limited seed dispersal is observed for either species (Calonje 2010). However, the longer generation times and more limited coning of sinkhole cycad would also act to reduce gene flow in a similar manner (cf. Kremer et al. 2012). The greater collection size of sinkhole cycad required for a fixed level of allele capture correlates with these reproductive biology factors. As a contrast to each Zamia in this study, the Keys thatch palm example examined an anemophilous, monoecious, panmictic species (Namoff et al. 2010; Griffith et al. 2011), and the much fewer numbers of ex situ plants required for high levels of genetic capture may correspond to these reproductive factors. Informed by these comparisons, we recommend that ex situ conservation plans include careful consideration of reproductive biology in sampling protocols.
Moving forward
By utilizing a comparison between two closely related species which differ in geographic and reproductive factors, we address some limitations about how broadly such findings can be applied. In this way, this study offers insight for ex situ collections protocols. Further parallel assays of other species, structured to include deliberate comparisons of rarity, reproductive isolation, dispersal, and generation times, would allow further testing of generalizations, and further refinement of protocols (Griffith and Husby 2010).
Adequate genetic capture is necessary for sustainable ex situ conservation collections, but sampling guidelines based on studies such as the current one must also take into account planned redundancy to mitigate against losses through reduced seed viability (Kay et al. 2011) and other losses (Griffith et al. 2008). A recent thread in the literature discusses ways in which collecting protocols insure against loss or drift (Guja et al. 2015; Guerrant et al. 2015; Hoban et al. 2015). Genetic capture via ex situ collections is only one portion of an integrated strategy for plant conservation (BGCI 2016), which can include basic research on plant diversity (Lorenzi et al. 2010), methods of sustainable use (Salomé-Castañeda et al. 2015), and involvement of stakeholders (García-Llorente et al. 2016).
Ex situ efforts currently maintain many plant species that would otherwise be extinct (Dhar 1996; Maunder et al. 2000; Sharrock 2011; Cousins et al. 2013). Despite these important cases, debate in the literature often questions the need for ex situ conservation (Hamilton 1994). Some root of this criticism is based on the perceived danger of authorities disregarding in situ conservation in favor of more politically feasible measures (Heywood 2009), or on inadequate attention to ecologically related issues (cf. Moir et al. 2012). Sometimes the critique of ex situ work is explicitly rooted in philosophical opinion (Rolston 2004). Careful review does note cases in which the benefits of ex situ conservation do not justify the costs (Clement et al. 2009). The conservation value of botanic garden ex situ collections can sometimes be overstated (Aplin 2008), most often due to insufficient data (Maunder et al. 2001).
We advance that the remedy for insufficient data is targeted data which allow rigorous assessment of conservation value. Studies such as the current work, which use targeted genetic data to assess the conservation value of ex situ collections, can help raise the understanding of exactly how well living collections can contribute to integrated efforts. Finally, cultivation of imperiled plants perfectly leverages the skills and assets of the botanic garden field to contribute to species survival. Regardless of the finer debate on the relative merits of ex situ botanic garden collections, the established feasibility of protective horticulture makes it an essential component in restoration efforts (Li and Pritchard 2009; Seaton et al. 2010; Vitt et al. 2010). Future world conditions may make ex situ work even more vital (Bridgewater 2016).
References
Aplin D (2008) How useful are botanic gardens for conservation? Plantsman 7:190–193
Arnet M, Santos B, Brockerhoff EG, Pelser PB, Ecroyd C, Clemens J (2015) Importance of arboreta for ex situ conservation of threatened trees. Biodivers Conserv 24:3601–3620
Berjak P, Pammenter NW (2014) Cryostorage of germplasm of tropical recalcitrant-seeded species: approaches and problems. Int J Plant Sci 175:29–39
BGCI (2014) Building living plant collections for conservation: a guide for public gardens. Montgomery Botanical Center. http://www.montgomerybotanical.org/media/Plant.Conservation.Collections.Guide.pdf. Accessed 3 Aug 2016
BGCI (2015) Cycads: a model group for ex situ plant conservation. Botanic Gardens Conservation International, Glencoe
BGCI (2016) North American Botanic Garden Strategy for Plant Conservation, 2016–2020. Botanic Gardens Conservation International, Glencoe
Bridgewater P (2016) The Anthropocene biosphere: do threatened species, red lists, and protected areas have a future role in nature conservation? Biodivers Conserv 25:603–607
Brown AHD (1989) Core collections: a practical approach to genetic resources management. Genome 31:818–824
Brütting C, Hensen I, Wesche K (2013) Ex situ cultivation affects genetic structure and diversity in arable plants. Plant Biology 15:505–513
Calonje M (2010) Bahamas cycad exploration begins. Montgomery Bot News 18:3
Calonje M, Husby C, Griffith P (2009a) The cycad collection at montgomery botanical center. Public Garden 24:25–27
Calonje M, Meerman J, Griffith P, Hoese G (2009b) A new species of Zamia (Zamiaceae) from the Maya Mountains of Belize. J Bot Res Inst Texas 31:31–41
Calonje C, Husby C, Calonje M (2010) Germination and early seedling growth of rare Zamia spp. in organic and inorganic substrates: advancing ex situ conservation horticulture. HortScience 45:679–683
Calonje M, Meerow AW, Knowles L, Knowles D, Griffith MP, Nakamura K, Francisco-Ortega J (2013) Cycad biodiversity in the Bahamas Archipelago and conservation genetics of the critically endangered Zamia lucayana (Zamiaceae). Oryx 47:190–198
Calonje M, Stevenson DW, Stanberg L (2016) The World List of Cycads, online edition. Montgomery Botanical Center, USA. http://www.cycadlist.org. Accessed 3 Aug 2016
Cavender N, Westwood M, Bechtoldt C, Donnelly G, Oldfield S, Gardner M, Rae D, McNamara W (2015) Strengthening the conservation value of ex situ tree collections. Oryx 49:416–424
Ceska JF, Affolter JM, Hamrick JL (1997) Developing a sampling strategy for Baptisia arachnifera based on allozyme diversity. Conserv Biol 11(5):1133–1139
Cibrian-Jaramillo A, Hird A, Oleas N, Ma H, Meerow AW, Francisco-Ortega J, Griffith MP (2013) What is the conservation value of a plant in a botanic garden? Using indicators to improve management of ex situ collections. Bot Rev 79:559–577
Clark DA, Clark DB (1987) Temporal and environmental patterns of reproduction in Zamia skinneri, a tropical rain forest cycad. J Ecol 75:135–149
Clement CR, Santos RP, Desmouliere SJM, Ferreira EJL, Neto JTF (2009) Ecological adaptation of wild peach palm, its in situ conservation and deforestation-mediated extinction in Southern Brazilian Amazonia. PLoS ONE 4:e4564. doi:10.1371/journal.pone.0004564
Clugston JAR, Griffith MP, Kenicer GJ, Husby CE, Calonje MA, Little DP, Stevenson DW (2017) Reproductive phenology of Zamia L.—a comparison between wild cycads and their cultivated counterparts. Memoirs of the New York Botanical Garden (in press)
Cohen J (1988) Statistical power analysis for the behavioral sciences. Routledge Academic, New York
Cousins SR, Williams VL, Witkowski ETF (2013) Sifting through cycads: a guide to identifying the stem fragments of six South African medicinal Encephalartos species. S Afr J Bot 84:115–123
Dhar S (1996) Corypha taliera: endangered palm extinct in the wild. Palm J 130:10–11
Duminil J, Fineschi S, Hampe A (2007) Can population genetic structure be predicted from life-history traits? Am Nat 169:662–672
Frankel OH (1984) Genetic perspectives of germplasm conservation. Genetic manipulation: impact on man and society. Cambridge University Press, Cambridge, pp 161–170
Frankham R (1995) Inbreeding and extinction: a threshold effect. Conserv Biol 9:792–799
Gale JS, Lawrence MJ (1984) The decay of variability. In: Holden JHW, Williams JT (eds) Crop genetic resources: conservation and evaluation. George Allen & Unwin, London
Gapare WJ, Aitken SN (2005) Strong spatial genetic structure in peripheral but not core populations of Sitka spruce [Picea sitchensis (Bong.) Carr.]. Mol Ecol 14:2659–2667
García-Llorente M., Harrison PA, Berry P, Palomo I, Gómez-Baggethun E, Iniesta-Arandia I, Montes C, del Amo DG, Martín-López B (2016) What can conservation strategies learn from the ecosystem services approach? Insights from ecosystem assessments in two Spanish protected areas. Biodivers Conserv. doi:10.1007/s10531-016-1152-4
Gemmill CEC, Ranker TA, Ragone D, Perlman SP, Wood KR (1998) Conservation genetics of the endangered endemic Hawaiian genus Brighamia (Campanulaceae). Am J Bot 85:528–539
Griffith P, Husby C (2010) The price of conservation: measuring the mission and its cost. BGJ 7:12–14
Griffith MP, Noblick LR, Dowe JL, Husby CE, Calonje M (2008) Cyclone tolerance in New World Arecaceae: biogeographic variation and abiotic natural selection. Ann Bot 102:591–598
Griffith P, Lewis C, Francisco-Ortega J (2011) Palm conservation in a botanic garden: a case study of the keys thatch palm. Palms 55:93–101
Griffith MP, Calonje MA, Stevenson DW, Husby CE, Little DP (2012) Time, place, and relationships: cycad phenology in a phylogenetic and biogeographic context. Mem NY Bot Garden 106:59–81
Griffith MP, Calonje M, Meerow AW, Tut F, Kramer AT, Hird A, Magellan TM, Husby CE (2015) Can a botanic garden cycad collection capture the genetic diversity in a wild population? Int J Plant Sci 176:1–10
Guerrant EO, Fiedler P, Havens K, Maunder M (2004) Revised genetic sampling guidelines for conservation collections of rare and endangered plants. In: Guerrant EO, Havens K, Maunder M (eds) Ex situ plant conservation: supporting species survival in the wild. Island Press, Washington, pp 419–441
Guerrant EO, Havens K, Vitt P (2014) Sampling for effective ex situ plant conservation. Int J Plant Sci 175:11–20
Guerrant EO, Havens K, Vitt P, Fiedler PL, Falk DA, Dixon K (2015) Population structure integral to seed collection guidelines: a response to Hoban and Schlarbaum (2014). Biol Cons 184:465–466
Guja LK, Broadhurst LM, Brown AH, Bush D, Cochrane A, Merritt DJ, Offord CA, Rossetto M, Wallace MJ, Wood J (2015) Genetic diversity is a significant but not the only consideration for effective ex situ plant conservation: response to Hoban and Schlarbaum. Biol Conserv 184:467–468
Haidet M, Olwell P (2015) Seeds of success: a national seed banking program working to achieve long-term conservation goals. Nat Areas J 35:165–173
Hamilton MB (1994) Ex situ conservation of wild plant species: time to reassess the genetic assumptions and implications of seed banks. Conserv Biol 8:39–49
Hamrick JL, Godt MJW (1996) Effects of life history traits on genetic diversity in plant species. Philos Trans Royal Soc B Biol Sci 351:1291–1298
Havens KA, Kramer AT, Guerrant EO (2014) Getting plant conservation right (or not): the case of the United States. Int J Plant Sci 175:3–10
Heywood V (2009) Botanic gardens and genetic conservation. Sibbaldia J Bot Garden Hortic 7:5–18
Hoban S, Schlarbaum S (2014) Optimal sampling of plant populations for ex situ conservation of genetic biodiversity, considering realistic population structure. Biol Conserv 177:90–99
Hoban S, Strand A (2015) Ex situ conservation seed collections should consider spatial design and species’ reproductive biology. Biol Conserv 187:182–191
Hoban S, Strand A, Fraga N, Richards C, Schlarbaum S (2015) Developing quantitative seed sampling protocols using simulations: a reply to comments from Guja et al and Guerrant et al. Biol Conserv 184:469–470
Hong TD, Ellis RH (1996) A protocol to determine seed storage behavior. IPGRI Technical Bulletin 1. International Plant Genetic Resources Institute, Rome
Huff DR, Peakall R, Smouse PE (1993) RAPD variation within and among natural populations of outcrossing buffalograss Buchloe dactyloides (Nutt) Engelm. Theor Appl Genet 86:927–934
Kay J, Strader A, Murphy V, Nghiem-Phu L, Calonje M, Griffith MP (2011) Palma Corcho: a case study in botanic garden conservation horticulture and economics. HortTechnology 21:474–481
Kremer A, Ronce O, Robledo-Arnuncio JJ, Guillaume F, Bohrer G, Nathan R, Bridle JR, Gomulkiewicz R, Klein EK, Ritland K, Kuparinen A (2012) Long-distance gene flow and adaptation of forest trees to rapid climate change. Ecol Lett 15:378–392
Krishnan S, Ranker TA, Davis AP, Rakotomalala JJ (2013) The study of genetic diversity patterns of Coffea commersoniana, an endangered coffee species from Madagascar: a model for conservation of other littoral forest species. Tree Genet Genomes 9:179–187
Larkin DJ, Jacobi SK, Hipp AL, Kramer AT (2016) Keeping All the PIECES: phylogenetically informed ex situ conservation of endangered species. PLoS ONE 11:e0156973
Li D-Z, Pritchard W (2009) The science and economics of ex situ plant conservation. Trends Plant Sci 14:614–621
Lopez-Gallego C, O’Neil P (2010) Life-history variation following habitat degradation associated with differing fine-scale spatial genetic structure in a rainforest cycad. Popul Ecol 52:191–201
Lorenzi H, Noblick LR, Kahn F, Ferreira E (2010) Brazilian flora Lorenzi: Arecaceae (Palms). Instituto Plantarum, Nova Odessa
Maunder M, Culham A, Alden B, Zizka G, Orliac C, Lobin W, Bordeu A, Ramirez JM, Glissmann-Gough S (2000) Conservation of the Toromiro tree: case study in the management of a plant extinct in the wild. Conserv Biol 14:1341–1350
Maunder M, Lyte B, Dransfield J, Baker W (2001) The conservation value of botanic garden palm collections. Biol Conserv 98:259–271
McGlaughlin ME, Riley L, Brandsrud M, Arcibal E, Helenurm MK, Helenurm K (2015) How much is enough? Minimum sampling intensity required to capture extant genetic diversity in ex situ seed collections: examples from the endangered plant Sibara filifolia (Brassicaceae). Conserv Genet 16:253–266
McGowen PJK, Traylor-Holzer K, Leus K (2016) IUCN Guidelines for determining when and how ex situ management should be used in species conservation. Conserv Lett 10:361–366
Meerow AW, Nakamura K (2007) Ten microsatellite loci from Zamia integrifolia (Zamiaceae). Mol Ecol Notes 7:824–826
Meerow AW, Francisco-Ortega J, Calonje M, Griffith MP, Ayala-Silva T, Stevenson DW, Nakamura K (2012) Zamia (Cycadales: Zamiaceae) on Puerto Rico: asymmetric genetic differentiation and the hypothesis of multiple introductions. Am J Bot 99:1828–1839
Menges ES, Guerrant EO, Hamze S (2004) Effects of seed collection on the extinction risk of perennial plants. In: Guerrant EO, Havens K, Maunder M (eds) Ex situ plant conservation: supporting species survival in the wild. Island, Washington, DC, pp 305–324
Moir ML, Vesk PA, Brennan KE, Poulin R, Hughes L, Keith DA, McCarthy MA, Coates DJ (2012) Considering extinction of dependent species during translocation, ex situ conservation, and assisted migration of threatened hosts. Conserv Biol 26:199–207
Namoff S, Husby CE, Francisco-Ortega J, Noblick LR, Lewis CE, Griffith MP (2010) How well does a botanical garden collection of a rare palm capture the genetic variation in a wild population? Biol Cons 143:1110–1117
Nei M (1978) Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics 89:583–590
O’Brien CW, Tang W (2015) Revision of the New World cycad weevils of the subtribe Allocorynina, with description of two new genera and three new subgenera (Coleoptera: Belidae: Oxycoryninae). Zootaxa 3970:1–187
Okubamichael DY, Jack S, Bösenberg JDW, Hoffman MT Donaldson JS (2016) Repeat photography confirms alarming decline in South African cycads. Biodivers Conserv
Orloci L (1978) Multivariate analysis in vegetation research, 2nd edn. Junk, The Hague
Pardey PG, Skovmand B, Taba S, Van Dusen ME, Wright BD (1998) The cost of conserving maize and wheat genetic resources ex situ. In: Smale M (ed) Farmers, gene banks and crop breeding: economic analyses of diversity in wheat maize and rice. Springer, Rotterdam, pp 35–56
Peakall R, Smouse PE (2006) GENALEX 6: genetic analysis in excel. Population genetic software for teaching and research. Mol Ecol Notes 6:288–295
Pence VC (2013) In vitro methods and the challenge of exceptional species for Target 8 of the Global Strategy for Plant Conservation. Ann Mo Bot Gard 99:214–220
Raven P, Havens K (2014) Ex situ plant conservation and cryopreservation: breakthroughs in tropical plant conservation. Int J Plant Sci 175:1–2
Richards CM, Antolin MF, Reilley A, Poole J, Walters C (2007) Capturing genetic diversity of wild populations for ex situ conservation: Texas wild rice (Zizania texana) as a model. Genet Resour Crop Evol 54:837–848
Rolston H III (2004) In Situ and Ex Situ Conservation: Philosophical and Ethical Concerns. In: Guerrant EO, Havens K, Maunder M (eds) Ex situ plant conservation: supporting species survival in the wild. Island, Washington, DC, pp 21–39
Salomé-Castañeda E, Mayett-Moreno Y, Barajas-Domínguez MI, Vovides AP (2015) Ornamental potential, sustainable use and some aspects of the marketing of Mexican cycads (Zamiaceae). Acta Hort 1104:511–516
Schemske DW, Husband BC, Ruckelshaus MH, Goodwillie C, Parker IM, Bishop JG (1994) Evaluating approaches to the conservation of rare and endangered plants. Ecology 75:584–606
Schoen DJ, Brown AHD (1991) Intraspecific variation in population gene diversity and effective population size correlates with the mating system in plants. Proc Natl Acad Sci 88(10):4494–4497
Schoen DJ, Brown AHD (2001) The conservation of wild plant species in seed banks. Bioscience 51:960–966
Seaton PT, Hu H, Perner H, Pritchard H (2010) Ex situ conservation of orchids in a warming world. Bot Rev 76:193–203
Sharrock SL (2011) The biodiversity benefits of botanic gardens. Trends Ecol Evol 26:433
Smith AB, Long QG, Albrecht MA (2016) Shifting targets: spatial priorities for ex situ plant conservation depend on interactions between current threats, climate change, and uncertainty. Biodivers Conserv 25:905–922
Touchell DH, Richardson M, Dixon KW (eds) (1997) Germplasm conservation guidelines for Australia: An introduction to the principles and practices for seed and germplasm banking of Australian species. Australian Network for Plant Conservation, Canberra
Vitt P, Havens K, Kramer AT, Sollenberger D, Yates E (2010) Assisted migration of plants: changes in latitudes, changes in attitudes. Biol Conserv 143:18–27
Volk GM, Richards CM, Reilley AA, Henk AD, Forsline PL, Aldwinckle HS (2005) Ex situ conservation of vegetatively propagated species: development of a seed-based core collection for Malus sieversii. J Am Soc Hortic Sci 130:203–210
Walters TW (2003) Off-Site Collections. In: Donaldson J (ed) Status survey and conservation action plan: Cycads. IUCN, Switzerland, pp 48–53
Walters T, Decker-Walters D (1991) Patterns of allozyme diversity in the West Indian cycad Zamia pumila (Zamiaceae). Am J Bot 78:436–449
Zaiontz C (2015) Real Statistics Using Excel. http://www.real-statistics.com
Acknowledgements
We thank the Ministry of the Environment of the Bahamas, Bahamas Department of Agriculture, Belize Forest Department, Belize Agricultural Health Authority, US Department of Agriculture and US Fish and Wildlife Service for permission to collect, study, and transport specimens. Research and analysis were generously funded by Institute of Museum and Library Services grants (MA-05-12-0336-12: Mission Based Collections Planning and MA-30-14-0123-14: Mission Based Collections Stewardship), marker development was funded by a National Science Foundation award (DEB 1050340), and living collections were supported by National Science Foundation awards (DBI 1203242 and DBI 1561346). Fieldwork was supported by the Association of Zoological Horticulture, SOS—Save Our Species (Grant 2012A-035), and the Mohamed bin Zayed Species Conservation Fund (Projects # 0925331 and # 12254271). The authors also thank these persons and organizations: Belize Defense Force, C. Calonje, S. Cuestas, J. duPlooy, duPlooy’s Jungle Lodge Resort, E. Freid, X. Gratacos, Green Hills Botanical Collections, A. Kramer, W. Mesh, J. Meerman, V. Murphy, K. Nakamura, D. Salas-Leiva, San Jose Hawaii Village, V. Tzub, and the Ya’axché Conservation Trust. This is contribution number 343 from the Tropical Biology Program of Florida International University.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by Daniel Sanchez Mata.
This article belongs to the Topical Collection: Ex-situ conservation.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Griffith, M.P., Calonje, M., Meerow, A.W. et al. Will the same ex situ protocols give similar results for closely related species?. Biodivers Conserv 26, 2951–2966 (2017). https://doi.org/10.1007/s10531-017-1400-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10531-017-1400-2