Abstract
Objective
To isolate putative lipase enzymes by screening a Cerrado soil metagenomic library with novel features.
Results
Of 6720 clones evaluated, Clone W (10,000 bp) presented lipolytic activity and four predicted coding sequences, one of them LipW. Characterization of a predicted esterase/lipase, LipW, showed 28% sequence identity with an arylesterase from Pseudomonas fluorescens (pdb|3HEA) from protein database (PDB). Phylogenetic analysis showed LipW clustered with family V lipases; however, LipW was clustered in different subclade belonged to family V, suggesting a different subgroup of family V. In addition, LipW presented a difference in family V GH motif, a glycine replaced by a serine in GH motif. Estimated molecular weight and stokes radius values of LipW were 29,338.67–29,411.98 Da and 2.58–2.83 nm, respectively. Optimal enzyme activity was observed at pH 9.0–9.5 and at 40 °C. Circular dichroism analysis estimated secondary structures percentages as approximately 45% α-helix and 15% β-sheet, consistent with the 3D structure predicted by homology.
Conclusion
Our results demonstrate the isolation of novel family V lipolytic enzyme with biotechnological applications from a metagenomic library.








Similar content being viewed by others
References
Adler AJ, Greenfield NJ, Fasman GD (1973) Circular dichroismus and optical rotatory dispersion of proteins and polypeptides. Methods Enzymol 27:675–735
Altschul S, Madden T, Schäffer A et al (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402
Arpigny JL, Jaeger KE (1999) Bacterial lipolytic enzymes: classification and properties. Biochem J 343(Pt 1):177–183
Arpigny JL, Jendrossek D, Jaeger KE (1998) A novel heat-stable lipolytic enzyme from Sulfolobus acidocaldarius DSM 639 displaying similarity to polyhydroxyalkanoate depolymerases. FEMS Microbiol Lett 167:69–73. https://doi.org/10.1016/S0378-1097(98)00375-9
Bairoch A, Boeckmann B (1994) The SWISS-PROT protein sequence data bank: current status. Nucleic Acids Res 22:3578–3580
Berman HM (2000) The Protein Data Bank. Nucleic Acids Res 28:235–242. https://doi.org/10.1093/nar/28.1.235
Bhattacharya D, Cheng J (2013) 3Drefine: consistent protein structure refinement by optimizing hydrogen bonding network and atomic-level energy minimization. Proteins 81:119–131. https://doi.org/10.1002/prot.24167
Böhm G, Muhr R, Jaeinicke R (1992) Quantitative analysis of protein far UV circular dichroism spectra by neural networks. Protein Eng 5:191–195
Bonelli FS, Jonas A (1993) Reaction of lecithin: cholesterol acyltransferase with a water soluble substrate: effects of surfactants. Biochim Biophys Acta 1166:92–98
Bornscheuer UT (2002) Microbial carboxyl esterases: classification, properties and application in biocatalysis. FEMS Microbiol Rev 26:73–81. https://doi.org/10.1016/S0168-6445(01)00075-4
Borrelli GM, Trono D (2015) Recombinant lipases and phospholipases and their use as biocatalysts for industrial applications. Int J Mol Sci 16:20774–20840. https://doi.org/10.3390/ijms160920774
Buchfink B, Xie C, Huson DH (2014) Fast and sensitive protein alignment using DIAMOND. Nat Methods 12:59–60. https://doi.org/10.1038/nmeth.3176
de Castro AP, Quirino BF, Allen H et al (2011) Construction and validation of two metagenomic DNA libraries from Cerrado soil with high clay content. Biotechnol Lett 33:2169–2175. https://doi.org/10.1007/s10529-011-0693-6
DeLano WL (2002) The PyMOL molecular graphics system. Schrödinger LLC wwwpymolorg, version 1. http://www.pymol.org
Delmont TO, Robe P, Cecillon S et al (2011) Accessing the soil metagenome for studies of microbial diversity. Appl Environ Microbiol 77:1315–1324. https://doi.org/10.1128/AEM.01526-10
Dewan SS (2017) Global markets for enzymes in industrial applications. www.bccresearch.com
dos Santos DFK, Istvan P, Noronha EF et al (2015) New dioxygenase from metagenomic library from Brazilian soil: insights into antibiotic resistance and bioremediation. Biotechnol Lett 37:1809–1817. https://doi.org/10.1007/s10529-015-1861-x
Dukunde A, Schneider D, Lu M et al (2017) A novel, versatile family IV carboxylesterase exhibits high stability and activity in a broad pH spectrum. Biotechnol Lett 39:577–587. https://doi.org/10.1007/s10529-016-2282-1
Feller G, Thiry M, Gerday C (1990) Sequence of a lipase gene from the antarctic psychrotroph Moraxella TA144. Nucleic Acids Res 18:6431. https://doi.org/10.1093/nar/18.21.6431
Ferrer M, Bargiela R, Martínez-Martínez M et al (2016) Biodiversity for biocatalysis: a review of the α/β-hydrolase fold superfamily of esterases-lipases discovered in metagenomes. Biocatal Biotransform 2422:1–15. https://doi.org/10.3109/10242422.2016.1151416
Gu X, Wang S, Wang S et al (2015) Identification and characterization of two novel Esterases from a metagenomic library. Food Sci Technol Res 21:649–657. https://doi.org/10.3136/fstr.21.649
Hide W, Chan A, Li WH (1992) Structure and evolution of the lipase superfamily. J Lipid Res 33:167–178
Hriscu M, Chiş L, Toşa M, Irimie FD (2013) pH-Profiling of thermoactive lipases and esterases: caveats and further notes. Eur J Lipid Sci Technol 115:571–575. https://doi.org/10.1002/ejlt.201200305
Hyatt D, Chen G-L, LoCascio PF et al (2010) Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinform 11:119. https://doi.org/10.1186/1471-2105-11-119
Jeon JH, Kim JT, Kang SG et al (2009) Characterization and its potential application of two esterases derived from the arctic sediment metagenome. Mar Biotechnol 11:307–316. https://doi.org/10.1007/s10126-008-9145-2
Katoh K, Standley DM (2013) MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30:772–780. https://doi.org/10.1093/molbev/mst010
Kumar S, Tsai C-J, Nussinov R (2000) Factors enhancing protein thermostability. Protein Eng Des Sel 13:179–191. https://doi.org/10.1093/protein/13.3.179
Larkin MA, Blackshields G, Brown NP et al (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948. https://doi.org/10.1093/bioinformatics/btm404
Lee MH, Lee S-W (2013) Bioprospecting potential of the soil metagenome: novel enzymes and bioactivities. Genom Inform 11:114. https://doi.org/10.5808/GI.2013.11.3.114
Lee MH, Khan R, Tao W et al (2018) Soil metagenome-derived 3-hydroxypalmitic acid methyl ester hydrolases suppress extracellular polysaccharide production in Ralstonia solanacearum. J Biotechnol 270:30–38. https://doi.org/10.1016/j.jbiotec.2018.01.023
Lenfant N, Hotelier T, Velluet E et al (2013) ESTHER, the database of the α/β-hydrolase fold superfamily of proteins: tools to explore diversity of functions. Nucleic Acids Res 41:423–429. https://doi.org/10.1093/nar/gks1154
Lovell SC, Davis IW, Arendall WB et al (2003) Structure validation by Calpha geometry: phi, psi and Cbeta deviation. Proteins 50:437–450. https://doi.org/10.1002/prot.10286
Newman JR, Fuqua C (1999) Broad-host-range expression vectors that carry the l-arabinose-inducible Escherichia coli araBAD promoter and the araC regulator. Gene 227:197–203. https://doi.org/10.1016/S0378-1119(98)00601-5
Notredame C, Higgins DG, Heringa J (2000) T-Coffee: a novel method for fast and accurate multiple sequence alignment. J Mol Biol 302:205–217. https://doi.org/10.1006/jmbi.2000.4042
Ovchinnikov S, Park H, Varghese N et al (2017) Protein structure determination using metagenome sequence data. Science 355:294–298. https://doi.org/10.1126/science.aah4043
Pereira MR, Mercaldi GF, Maester TC et al (2015) Est16, a new esterase isolated from a metagenomic library of a microbial consortium specializing in diesel oil degradation. PLoS ONE 10:1–16. https://doi.org/10.1371/journal.pone.0133723
Price MN, Dehal PS, Arkin AP (2010) FastTree 2 - Approximately maximum-likelihood trees for large alignments. PLoS ONE. https://doi.org/10.1371/journal.pone.0009490
Robert X, Gouet P (2014) Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res 42:W320–W324. https://doi.org/10.1093/nar/gku316
Schuck P (2000) Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and Lamm equation modeling. Biophys J 78:1606–1619. https://doi.org/10.1016/S0006-3495(00)76713-0
Schuck P (2003) On the analysis of protein self-association by sedimentation velocity analytical ultracentrifugation. Anal Biochem 320:104–124. https://doi.org/10.1016/S0003-2697(03)00289-6
Torsvik V, Goksoyr J, Daae FL (1990) High diversity in DNA of soil bacteria. Appl Environ Microbiol 56:782–787. https://doi.org/10.1017/CBO9781107415324.004
Tyzack JD, Furnham N, Sillitoe I et al (2017) Understanding enzyme function evolution from a computational perspective. Curr Opin Struct Biol 47:131–139. https://doi.org/10.1016/j.sbi.2017.08.003
Valle A, Pérez-Socas LB, Canet L et al (2018) Self-homodimerization of an actinoporin by disulfide bridging reveals implications for their structure and pore formation. Sci Rep 8:1–18. https://doi.org/10.1038/s41598-018-24688-2
Webb B, Sali A (2014) Protein structure modeling with MODELLER. Methods Mol Biol 1137:1–15. https://doi.org/10.1007/978-1-4939-0366-5_1
Xing MN, Zhang XZ, Huang H (2012) Application of metagenomic techniques in mining enzymes from microbial communities for biofuel synthesis. Biotechnol Adv 30:920–929. https://doi.org/10.1016/j.biotechadv.2012.01.021
Yang X, Wu L, Xu Y et al (2018) Identification and characterization of a novel alkalistable and salt-tolerant esterase from the deep-sea hydrothermal vent of the East Pacific Rise. Microbiologyopen. https://doi.org/10.1002/mbo3.601
Yin DL, Bernhardt P, Morley KL et al (2010) Switching Catalysis from Hydrolysis to Perhydrolysis in Pseudomonas fluorescens Esterase. Biochemistry 49:1931–1942. https://doi.org/10.1021/bi9021268
Acknowledgments
The authors thank National Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) and Fundação de Apoio à Pesquisa do Distrito Federal (FAP-DF) for financial support. Istvan and Lopes acknowledge a fellowship from Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES).
Supporting information
Online Resource 1—Annotation and taxonomic classification of the predicted coding regions of Clone W. The translated sequences of predicted CDSs (> 150 aa) were aligned with protein sequences in the SwissProt, TrEMBL, and UniRef100 databases. Only the top hits of each alignment are shown.
Online Resource 2—Sedimentation velocity. a Raw data of absorbance at 280 nm versus cell radius for 2.91 μM LipW. b Residuals plot supplied by SEDFIT showing goodness of fit. (c) Continuous sedimentation coefficient distribution, c(S) curve, obtained with a regularization procedure from data shown in panel a with a confidence level of 0.95 using SEDFIT and frictional ratios of 1.20–1.31. The partial specific volume (υ) 0.735614 ml/g for LipW was determined using SEDNTERP. Solvent (water) density (ρ = 0.99823 g/ml) and viscosity (η = 0.01002 poise) were also determined by SEDNTERP. Circles represent experimental data, and the solid line represents the best fit to the Lamm equation supplied by SEDFIT. Similar results were obtained for 2.14 and 1.56 μM LipW.
Online Resource 3—Molecular and biophysical parameters of LipW.
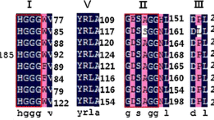
Online Resource 4—Amino acid sequence alignment of LipW with family V lipases. The G-X-S-M-G–G consensus motif of family V is shown, and the catalytic triad residues are denoted with asterisks.
Online Resource 5—Ramachandran plot of the LipW model.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Istvan, P., Souza, A.A., Garay, A.V. et al. Structural and functional characterization of a novel lipolytic enzyme from a Brazilian Cerrado soil metagenomic library. Biotechnol Lett 40, 1395–1406 (2018). https://doi.org/10.1007/s10529-018-2598-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10529-018-2598-0