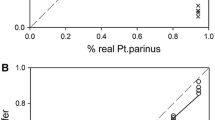
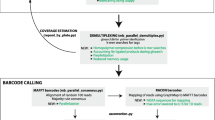
Abstract
Feather mites are among the most common and diverse ectosymbionts of birds, yet basic questions such as the nature of their relationship remain largely unanswered. One reason for feather mites being understudied is that their morphological identification is often virtually impossible when using female or young individuals. Even for adult male specimens this task is tedious and requires advanced taxonomic expertise, thus hampering large-scale studies. In addition, molecular-based methods are challenging because the low DNA amounts usually obtained from these tiny mites do not reach the levels required for high-throughput sequencing. This work aims to overcome these issues by using a DNA metabarcoding approach to accurately identify and quantify the feather mite species present in a sample. DNA metabarcoding is a widely used molecular technique that takes advantage of high-throughput sequencing methodologies to assign the taxonomic identity to all the organisms present in a complex sample (i.e., a sample made up of multiple specimens that are hard or impossible to individualise). We present a high-throughput method for feather mite identification using a fragment of the COI gene as marker and Illumina Miseq technology. We tested this method by performing two experiments plus a field test over a total of 11,861 individual mites (5360 of which were also morphologically identified). In the first experiment, we tested the probability of detecting a single feather mite in a heterogeneous pool of non-conspecific individuals. In the second experiment, we made 2 × 2 combinations of species and studied the relationship between the proportion of individuals of a given species in a sample and the proportion of sequences retrieved to test whether DNA metabarcoding can reliably quantify the relative abundance of mites in a sample. Here we also tested the efficacy of degenerate primers (i.e., a mixture of similar primers that differ in one or several bases that are designed to increase the chance of annealing) and investigated the relationship between the number of mismatches and PCR success. Finally, we applied our DNA metabarcoding pipeline to a total of 6501 unidentified and unsorted feather mite individuals sampled from 380 European passerine birds belonging to 10 bird species (field test). Our results show that this proposed pipeline is suitable for correct identification and quantitative estimation of the relative abundance of feather mite species in complex samples, especially when dealing with a moderate number (> 30) of individuals per sample.



Similar content being viewed by others
Data accessibility
The MiSeq raw data, the processed representative sequences files, and the custom-made bioinformatic scripts have been deposited in Figshare (https://doi.org/10.6084/m9.figshare.5527405).
References
Allen JM, Boyd B, Nguyen NP et al (2017) Phylogenomics from whole genome sequences using aTRAM. Syst Biol 66:786–798
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215:403–410
Andrew S (2010) FastQC, a quality control tool for high throughput sequence data. Retrieved Oct 2015, from http://www.bioinformatics.babraham.ac.uk/projects/fastqc
Arribas P, Andujar C, Hopkins K, Shepherd M, Vogler AP (2016) Metabarcoding and mitochondrial metagenomics of endogean arthropods to unveil the mesofauna of the soil. Methods Ecol Evol 7:1071–1081
Atyeo WT, Braasch NL (1966) The feather mite genus Proctophyllodes (Sarcoptiformes: Proctophyllodidae). Univ Neb State Mus 5:1–354
Atyeo WT, Gaud J (1970) The feather mite genus Monojourbertia Radford, 1950 (Analgoidea: Proctophyllodidae). Entomologische Mitteilungen aus dem Zoologischen Staatsinstitut und Zoologischen Museum, Hamburg 4:145–155
Baker CC, Bittleston LS, Sanders JG, Pierce NE (2016) Dissecting host-associated communities with DNA barcodes. Philos Trans R Soc B 371:20150328
Blanco G, Tella J, Potti J, Baz A (2001) Feather mites on birds: costs of parasitism or conditional outcomes? J Avian Biol 32:271–274
Bolker BM (2008) Ecological models and data in R. Princeton University Press, Princeton
Bushnell B (2014) BBMap: a fast, accurate, splice-aware aligner. Report number: LBNL-7065E, Lawrence Berkeley National Laboratory, Berkeley, CA
Caporaso JG, Kuczynski J, Stombaugh J et al (2010) QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7:335–336
Carlsen T, Aas AB, Lindner D, Vrålstad T, Schumacher T, Kauserud H (2012) Don’t make a mista(g)ke: is tag switching an overlooked source of error in amplicon pyrosequencing studies? Fungal Ecol 5:747–749
Carlson CJ, Burgio KR, Dougherty ER et al (2017) Parasite biodiversity faces extinction and redistribution in a changing climate. Sci Adv 3:e1602422
Dabert J, Ehrnsberger R, Dabert M (2008) Glaucalges tytonis sp. nov. (Analgoidea: Xolalgidae) from the barn owl Tyto alba (Strigiformes: Tytonidae): compiling morphology with DNA barcode data for taxa descriptions in mites (Acari). Zootaxa 1719:41–52
De Tender CA, Devriese LI, Haegeman A, Maes S, Ruttink T, Dawyndt P (2015) Bacterial community profiling of plastic litter in the Belgian part of the North Sea. Environ Sci Technol 49:9629–9638
Deagle BE, Jarman SN, Coissac E, Pompanon F, Taberlet P (2014) DNA metabarcoding and the cytochrome c oxidase subunit I marker: not a perfect match. Biol Lett 10:20140562
Diaz-Real J, Serrano D, Pérez-Tris J et al (2014) Repeatability of feather mite prevalence and intensity in passerine birds. PLoS ONE 9:e107341
Diaz-Real J, Serrano D, Piriz A, Jovani R (2015) NGS metabarcoding proves successful for quantitative assessment of symbiont abundance: the case of feather mites on birds. Exp Appl Acarol 67:209–218
Dobson A, Lafferty K, Kuris A, Hechinger R, Jetz W (2008) Homage to Linnaeus: how many parasites? How many hosts? Proc Natl Acad Sci 105:11482–11489
Doña J, Diaz-Real J, Mironov S, Bazaga P, Serrano D, Jovani R (2015a) DNA barcoding and mini-barcoding as a powerful tool for feather mite studies. Mol Ecol Resour 15:1216–1225
Doña J, Moreno-García M, Criscione CD, Serrano D, Jovani R (2015b) Species mtDNA genetic diversity explained by infrapopulation size in a host-symbiont system. Ecol Evolut 5:5801–5809
Doña J, Proctor H, Mironov S, Serrano D, Jovani R (2016) Global associations between birds and vane-dwelling feather mites. Ecology 97:3242
Doña J, Potti J, De la Hera I, Blanco G, Frías O, Jovani R (2017a) Vertical transmission in feather mites: insights into its adaptive value. Ecol Entomol 42:492–499
Doña J, Sweet AD, Johnson KP, Serrano D, Mironov S, Jovani R (2017b) Cophylogenetic analyses reveal extensive host-shift speciation in a highly specialized and host-specific symbiont system. Mol Phylogenet Evol 115:190–196
Doña J, Proctor H, Serrano D et al (2018) Feather mites play a role in cleaning host feathers: new insights from DNA metabarcoding and microscopy. Mol Ecol. https://doi.org/10.1111/mec.14581
Dubinin VB (1951) Feather mites (Analgesoidea). Part 1. Introduction to their study. Fauna USSR 6:1–363 (in Russian)
Edgar RC (2010) Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26:2460–2461. https://doi.org/10.1093/bioinformatics/btq461
Elbrecht V, Leese F (2017) PrimerMiner: an R package for development and in silico validation of DNA metabarcoding primers. Methods Ecol Evol 8:622–626
Elbrecht V, Vamos EE, Meissner K, Aroviita J, Leese F (2017) Assessing strengths and weaknesses of DNA metabarcoding-based macroinvertebrate identification for routine stream monitoring. Methods Ecol Evol 8:1265–1275
Esling P, Lejzerowicz F, Pawlowski J (2015) Accurate multiplexing and filtering for high-throughput amplicon-sequencing. Nucl Acids Res 43:2513–2524
Ferrari S, Cribari-Neto F (2004) Beta regression for modelling rates and proportions. J Appl Stat 31:799–815
Ficetola G, Pansu J, Bonin A et al (2015) Replication levels, false presences and the estimation of the presence/absence from eDNA metabarcoding data. Mol Ecol Resour 15:543–556
Gaud J, Atyeo WT (1996) Feather mites of the World (Acarina, Astigmata): the supraspecific taxa. Annales du Musee Royale de L’Afrique Centrale, Sciences Zoologiques, 277, 1–193 (Pt. 1, text), 1–436 (Pt. 2, illustrations)
Geisen S, Laros I, Vizcaíno A, Bonkowski M, de Groot GA (2015) Not all are free-living: high-throughput DNA metabarcoding reveals a diverse community of protists parasitizing soil metazoa. Mol Ecol 24:4556–4569
Hawkins TL, O’Connor-Morin T, Roy A, Santillan C (1994) DNA purification and isolation using a solid-phase. Nucl Acids Res 22:4543–4544
Hebert PDN, Cywinska A, Ball SL, deWaard JR (2003) Biological identifications through DNA barcodes. Proc R Soc Lond B 270:313–321
Jousselin E, Clamens AL, Galan M et al (2016) Assessment of a 16S rRNA amplicon illumina sequencing procedure for studying the microbiome of a symbiont-rich aphid genus. Mol Ecol Resour 16:628–640
Kearse M, Moir R, Wilson A et al (2012) Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28:1647–1649
Lafferty KD, Dobson A, Kuris AM (2006) Parasites dominate food web links. Proc Natl Acad Sci 103:11211–11216
Lange V, Böhme I, Hofman J et al (2014) Cost-efficient high-throughput HLA typing by MiSeq amplicon sequencing. BMC Genom 15:63
Linard B, Arribas P, Andújar C, Crampton-Platt A, Vogler AP (2016) Lessons from genome skimming of arthropod-preserving ethanol. Mol Ecol Resour 16:1365–1377
Meléndez L, Laiolo P, Mironov S, García M, Magaña O, Jovani R (2014) Climate-driven variation in the intensity of a host-symbiont animal interaction along a broad elevation gradient. PLoS ONE 9:e101942
Mironov SV, Galloway TD (2006) New and little-known species of the feather mites (Acari: Analgoidea: Pteronyssidae) from birds in North America. Can Entomol 138:165–188
Mironov SV, Wauthy G (2006) Systematic review of feather mites of the genus Sturnotrogus Mironov, 1989 (Astigmata: Pteronyssidae) from starlings (Passeriformes: Sturnidae) in Africa and Europe. Bulletin de l’Institut Royal des Sciences naturelles de Belgique, Entomogie 76:55–81
Mironov SV, Dabert J, Dabert M (2012) A new feather mite species of the genus Proctophyllodes Robin, 1877 (Astigmata: Proctophyllodidae) from the Long-tailed Tit Aegithalos caudatus (Passeriformes: Aegithalidae)—morphological description with DNA barcode data. Zootaxa 3253:54–61
Mironov SV, Doña J, Jovani R (2015) A new feather mite of the genus Dolichodectes (Astigmata: Proctophyllodidae) from Hippolais polyglotta (Passeriformes: Acrocephalidae) in Spain. Folia Parasitol 62:032
Navarro-Noya YE, Valenzuela-Encinas C, Sandoval-Yuriar A, Jiménez-Bueno NG, Marsch R, Dendooven L (2015) Archaeal communities in a heterogeneous hypersaline-alkaline soil. Archaea 2015:11
Owens GL, Todesco M, Drummond EB, Yeaman S, Rieseberg LH (2018) A novel post hoc method for detecting index switching finds no evidence for increased switching on the Illumina HiSeq X. Mol Ecol Resour 18:169–175
Pap P, Vágási C, Osváth G, Mureşan C, Barta Z (2010) Seasonality in the uropygial gland size and feather mite abundance in house sparrows Passer domesticus: natural covariation and an experiment. J Avian Biol 41:653–661
Park CK, Atyeo WT (1971) A generic revision of the Pterodectinae, a new subfamily of feather mites (Sarcoptiformes: Analgoidea). Bull Univ Neb State Mus 9:39–88
Pornon A, Escaravage N, Burrus M et al (2016) Using metabarcoding to reveal and quantify plant–pollinator interactions. Sci Rep 6:27282
Poulin R (2014) Parasite biodiversity revisited: frontiers and constraints. Int J Parasitol 44:581–589
Proctor H (2003) Feather mites (Acari: Astigmata): ecology, behavior, and evolution. Annu Rev Entomol 48:185–209
Reva ON, Zaets IE, Ovcharenko LP et al (2015) Metabarcoding of the kombucha microbial community grown in different microenvironments. AMB Express 5:124
Riaz T, Shehzad W, Viari A, Pompanon F, Taberlet P, Coissac E (2011) ecoPrimers: inference of new DNA barcode markers from whole genome sequence analysis. Nucl Acids Res 39:e145–e145
Rocha CFD, Bergallo HG, Bittencourt EB (2016) More than just invisible inhabitants: parasites are important but neglected components of the biodiversity. Zoologia (Curitiba) 33:e20150198
R Development Core Team (2015) R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. ISBN 3-900051-07-0. http://www.R-project.org/. Accessed July 2017
Santana F (1976) A review of the genus Trouessartia: (Analgoidea: Alloptidae). J Med Entomol 13:1–125
Schnell IB, Bohmann K, Gilbert MT (2015) Tag jumps illuminated: reducing sequence-to-sample misidentifications in metabarcoding studies. Mol Ecol Resour 15:1289–1303
Schrader C, Schielke A, Ellerbroek L, Johne R (2012) PCR inhibitors: occurrence, properties and removal. J Appl Microbiol 113:1014–1026
Sinha R, Stanley G, Gulati G et al (2017) Index switching causes “spreading-of-signal” among multiplexed samples in Illumina HiSeq 4000 DNA sequencing. bioRxiv. https://doi.org/10.1101/125724
Sipos R, Székely A, Palatinszky M, Révész S, Márialigeti K, Nikolausz M (2007) Effect of primer mismatch, annealing temperature and PCR cycle number on 16S rRNA gene-targetting bacterial community analysis. FEMS Microbiol Ecol 60:341–350
Soininen EM, Zinger L, Gielly L et al (2013) Shedding new light on the diet of Norwegian lemmings: DNA metabarcoding of stomach content. Polar Biol 36:1069–1076
Stephens ZD, Lee SY, Faghri F et al (2015) Big data: astronomical or genomical? PLoS Biol 13:1–11
Taberlet P, Coissac E, Hajibabaei M, Rieseberg LH (2012a) Environmental DNA. Mol Ecol 21:1789–1793
Taberlet P, Coissac E, Pompanon F, Brochmann C, Willerslev E (2012b) Towards next-generation biodiversity assessment using DNA metabarcoding. Mol Ecol 21:2045–2050
Thomas CD, Cameron A, Green RE et al (2004) Extinction risk from climate change. Nature 427:145–148
Tripp E, Zhang N, Schneider H et al (2017) Reshaping Darwin’s tree: impact of the symbiome. Trends Ecol Evol 32:552–555
Truett GE, Heeger P, Mynatt RL, Truett AA, Walker JA, Warman ML (2000) Preparation of PCR-quality mouse genomic DNA with hot sodium hydroxide and tris (HotSHOT). Biotechniques 29:52–54
Vierna J, Doña J, Vizcaíno A, Serrano D, Jovani R (2017) PCR cycles above routine numbers do not compromise high-throughput DNA barcoding results. Genome 60:868–873
Wang Q, Garrity GM, Tiedje JM, Cole JR (2007) Naive bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 73:5261–5267
Wolak ME, Fairbairn DJ, Paulsen YR (2012) Guidelines for estimating repeatability. Methods Ecol Evol 3:129–137
Zeileis A, Cribari-Neto F, Gruen B, Kosmidis I (2012) Package ‘betareg’. https://cran.r-project.org/web/packages/betareg/betareg.pdf
Acknowledgements
Funding was provided by the Spanish Ministry of Economy and Competitiveness (Ramón y Cajal research contract RYC-2009-03967 to RJ, research project CGL2011-24466 to RJ, and CGL2015-69650-P to RJ and DS). JD was supported by the Spanish Ministry of Economy and Competitiveness (Severo Ochoa predoctoral contract SVP-2013-067939), and SV was supported by the Russian Foundation for Basic Research (RFBR-6-04-00486). Special thanks for the help in collecting samples to: Carolina Osuna, Alberto Álvarez, Emilio Paganí-Núñez, Carlos Gutiérrez Expósito, Carlos Camacho, David Ochoa, Jaime Potti, José Luis Arroyo, Rubén Rodríguez Olivares, Marina Moreno-García, Pepe Ayala, Jose Luis Garzón, Francisco Jimenez Cazalla and Sociedad Ornitológica de Menorca (SOM).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Vizcaíno, A., Doña, J., Vierna, J. et al. Enabling large-scale feather mite studies: an Illumina DNA metabarcoding pipeline. Exp Appl Acarol 76, 81–97 (2018). https://doi.org/10.1007/s10493-018-0288-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10493-018-0288-1