Abstract
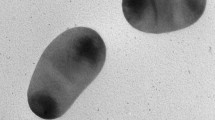
A Gram-negative, rod shaped, motile bacterium, was isolated from a marine solar saltern sample collected from Kakinada, India. Strain AK2T was determined to be positive for nitrate reduction, catalase, Ala-Phe-Pro-arylamidase, β-galactosidase, β-N-acetylglucosaminidase, β-glucosidase, β-xylosidase, α-glucosidase, α-galactosidase and phosphatase activities, hydrolysis of aesculin, Tween 20/40/60/80 and urea. It was determined to be negative for oxidase, lysine decarboxylase and ornithine decarboxylase activities and could not hydrolyze agar, casein, gelatin and starch. The predominant fatty acids were identified as iso-C15:0 (28.2 %), anteiso-C15:0 (23.2 %), iso-C13:0 (19.9 %) and iso-C15:0 3-OH (13.9 %). Strain AK2T was found to contain menaquinone with seven isoprene units (MK-7) as the sole respiratory quinone and phosphatidylethanolamine, one unidentified phospholipid and three unidentified lipids as polar lipids. The 16S rRNA gene sequence analysis indicated the strain AK2T as a member of the genus Marinilabilia and is closely related to Marinilabilia salmonicolor with pair-wise sequence similarity of 98.2 %. Phylogenetic analysis of 16S rRNA gene revealed that the strain AK2T clustered with M. salmonicolor. However, DNA–DNA hybridization with M. salmonicolor JCM 21150T showed a relatedness of 48 ± 0.5 % with respect to strain AK2T. The DNA G+C content of the strain was determined to be 40.2 mol%. Based on the phenotypic characteristics and phylogenetic inference, it is proposed that the strain AK2T represents a novel species of the genus Marinilabilia, for which the name Marinilabilia nitratireducens sp. nov. is proposed. The type strain of M. nitratireducens sp. nov. is AK2T (= MTCC 11402T = JCM 17679T).


Similar content being viewed by others
References
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. Mol Biol 215:403–410
Anil Kumar P, Srinivas TNR, Madhu S, Ruth Manorama R, Shivaji S (2010) Indibacter alkaliphilus gen. nov., sp. nov., a novel alkaliphilic bacterium isolated from haloalkaline Lonar Lake, India. Int J Syst Evol Microbiol 60:721–726
Bernardet JF, Nakagawa Y, Holmes B (2002) Subcommittee on the taxonomy of Flavobacterium and Cytophaga-like bacteria of the International Committee on Systematics of Prokaryotes. Proposed minimal standards for describing new taxa of the family Flavobacteriaceae and emended description of the family. Int J Syst Evol Microbiol 52:1049–1070
Bligh EG, Dyer WJ (1959) A rapid method of total lipid extraction and purification. Can J Biochem Physiol 37:911–917
Collins MD, Pirouz T, Goodfellow M, Minnikin DE (1977) Distribution of menaquinones in actinomycetes and corynebacteria. Gen Microbiol 100:221–230
Groth I, Schumann P, Rainey FA, Martin K, Schuetze B, Augsten K (1997) Demetria terragena gen. nov., sp. nov., a new genus of actinomycetes isolated from compost soil. Int J Syst Bacteriol 47:1129–1133
Johnson JL (1994) Similarity analysis of rRNAs. In: Gerhardt P, Murray RGE, Wood WA, Krieg NR (eds) Methods for General and Molecular Bacteriology. American Society for Microbiology, Washington, pp 683–700
Komagata K, Suzuki K (1987) Lipid and cell wall analysis in bacterial systematics. Methods Microbiol 19:161–206
Lányí B (1987) Classical and rapid identification methods for medically important bacteria. Methods Microbiol 19:1–67
Ludwig W, Euzéby J, Whitman WB (2012) Family II. Marinilabiliaceae fam. nov. In: Krieg NR, Staley JT, Brown DR, Hedlund BP, Paster BJ, Ward NL, Ludwig W, Whitman WB (eds) Bergey’s manual of systematic bacteriology (The Bacteroidetes, Spirochaetes, Tenericutes (Mollicutes), Acidobacteria, Fibrobacteres, Fusobacteria, Dictyoglomi, Gemmatimonadetes, Lentisphaerae, Verrucomicrobia, Chlamydiae, and Planctomycetes), vol 4, 2nd edn. Springer, New York, p 49
Marmur J (1961) A procedure for the isolation of deoxyribonucleic acid from microorganisms. Mol Biol 3:208–218
Miyazaki M, Koide O, Kobayashi T, Mori K, Shimamura S, Nunoura T, Imachi H, Inagaki F, Nagahama T, Nogi Y, Deguchi S, Takai K (2011) Geofilum rubicundum gen. nov. sp. nov., isolated from deep subseafloor sediment. Int J Syst Evol Microbiol 62:1075–1080
Nakagawa Y (2011) Class IV. Cytophagia class. nov. In: Krieg NR, Staley JT, Brown DR, Hedlund BP, Paster BJ, Ward NL, Ludwig W, Whitman WB (eds) Bergey’s manual of systematic bacteriology (The Bacteroidetes, Spirochaetes, Tenericutes (Mollicutes), Acidobacteria, Fibrobacteres, Fusobacteria, Dictyoglomi, Gemmatimonadetes, Lentisphaerae, Verrucomicrobia, Chlamydiae, and Planctomycetes), vol 4, 2nd edn. Springer, New York, p 370
Nakagawa Y, Yamasato K (1996) Emendation of the genus Cytophaga and transfer of Cytophaga agarovorans and Cytophaga salmonicolor to Marinilabilia gen. nov.: phylogenetic analysis of the Marinilabilia-Cytophaga complex. Int J Syst Bacteriol 46:599–603
Nei M, Kumar S (2000) Molecular Evolution and Phylogenetics. Oxford University Press, New York
Pandey KK, Mayilraj S, Chakrabarti T (2002) Pseudomonas indica sp. nov., a novel butane-utilizing species. Int J Syst Evol Microbiol 52:1559–1567
Reichenbach H (1989) Genus 1. Cytophagu. In: Staley JT, Bryant MP, Pfennig N, Holt JG (eds) Bergey’s manual of systematic bacteriology, vol 3. The Williams and Wilkins Co., Baltimore, pp 2015–2050
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Sly LI, Blackall LL, Kraat PC, Tao T-S, Sangkhobol V (1986) The use of second derivative plots for the determination of mol% guanine plus cytosine of DNA by the thermal denaturation method. Microbiol Methods 5:139–156
Smibert RM, Krieg NR (1994) Phenotypic characterization. In: Gerhardt P, Murray RGE, Wood WA, Krieg NR (eds) Methods for General and Molecular Bacteriology. American Society for Microbiology, Washington, pp 607–654
Srinivas TNR, Anil Kumar P, Madhu S, Sunil B, Sharma TVRS, Shivaji S (2011) Cesiribacter andamanensis gen. nov., sp. nov., a novel bacterium isolated from a soil sample of mud volcano, Andaman Islands India. Int J Syst Evol Microbiol 61:1521–1527
Surendra V, Pant B, Korpole S, Srinivas TNR, Anil Kumar P. (2011) Imtechella halotolerans gen. nov., sp. nov., a member of the family Flavobacteriaceae isolated from estuarine water. Int J Syst Evol Microbiol (In Press) doi:10.1099/ijs.0.038356-0
Suzuki M, Nakagawa Y, Harayama S, Yamamoto S (1999) Phylogenetic analysis of genus Marinilabilia and related bacteria based on the amino acid sequences of GyrB and emended description of Marinilabilia salmonicolor with Marinilabilia agarovorans as its subjective synonym. Int J Syst Bacteriol 49:1551–1557
Tamura K, Nei M (1993) Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol 10:512–526
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. doi:10.1093/molbev/msr121
Tourova TP, Antonov AS (1987) Identification of microorganisms by rapid DNA–DNA hybridisation. Methods Microbiol 19:333–355
Veldkamp H (1961) A study of two marine agar-decomposing, facultatively anaerobic myxobacteria. J Gen Microbiol 26:331–342
Wayne LG, Brenner DJ, Colwell RR, Grimont PAD, Kandler O, Krichevsky MI, Moore LH, Moore WEC, Murray RGE et al (1987) Report of the ad hoc committee on reconciliation of approaches to bacterial systematics. Int J Syst Bacteriol 37:463–464
Acknowledgments
We thank Dr. J. Euzeby for his expert suggestion for the correct species epithet and Latin etymology. We also thank Council of Scientific and Industrial Research (CSIR) and Department of Biotechnology, Government of India for financial assistance. We would like to thank Mr. Deepak for his help in sequencing DNA. TNRS is thankful to CSIR OLP project (OLP1201) for providing facilities and funding respectively. NIO contribution number is 5250.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Shalley, S., Pradip Kumar, S., Srinivas, T.N.R. et al. Marinilabilia nitratireducens sp. nov., a lipolytic bacterium of the family Marinilabiliaceae isolated from marine solar saltern. Antonie van Leeuwenhoek 103, 519–525 (2013). https://doi.org/10.1007/s10482-012-9834-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10482-012-9834-8