Abstract
Although the biogeographic history of thrushes (Turdidae) has been extensively studied, a concise discussion of this topic is still lacking. Therefore, in this study we aimed to investigate: (1) the evolutionary origin of the migratory behaviour of the Turdus thrushes in a biogeographic context including (2) trans-Atlantic dispersal events, (3) possible colonization routes into the Nearctic, and (4) relationships among life history traits, ecological factors, and migratory strategies within the most comprehensive taxon set of 72 Turdus thrushes to date. We estimated the ancestral ranges of the studied species, primarily by comparing main biogeographic models (dispersal-vicariance, dispersal-extinction-cladogenesis, BayArea models), and performed phylogenetic generalized least squares analyses to identify relationships among distribution patterns, diet, body measurements, clutch size, and migratory behaviour. We found that the most probable ancestral regions for all Turdus species were located in the East Palearctic realm, followed by early colonization of the western Palearctic and Africa, and that several trans-Atlantic movements occurred between 11 and 4 million years ago, which is earlier than previously thought. Migration emerged as an ancestral behaviour of the genus Turdus, and differences in clutch size and main food types were significant between migratory and non-migratory species. Correlated evolution was found between migration and along-latitudinal mobility, main food type, and the shift in main food type between the seasons. We conclude that along-latitudinal movements may have evolved earlier, simultaneously with the radiation of the Turdus thrushes, followed by the appearance of meridional migration, associated with orographic and climatic changes. The increased clutch sizes observed in migratory species and documented here for thrushes could serve as an important mechanism to compensate for losses due to mortality during migration.
Zusammenfassung
Phylogenie, Zugverhalten, Lebensgeschichte: ein Versuch, Lücken in der Biogeographie und Herkunft der Turdus -Drosseln zu schließen.
Obwohl die biogeographische Geschichte der Drosselartigen (Turdidae) schon umfassend untersucht wurde, fehlt jedoch eine umfassende Diskussion zu diesem Thema. Deswegen wollten wir in dieser Studie untersuchen: (1) den evolutionären Ursprung des Zugverhaltens in einem biogeographischen Kontext, einschließlich, (2) transatlantische Ausbreitungsereignisse, (3) mögliche Kolonisationswege in die Nearktis und (4) Beziehungen zwischen lebensgeschichtlichen Merkmalen, ökologischen Faktoren und Zugstrategien innerhalb des bisher umfassendsten Taxonbestandes von 72 Turdus-Drosseln. Wir haben die Herkunftsareale der untersuchten Arten berechnet, vor allem durch den Vergleich der hauptsächlichen biogeographischen Modelle (DIVA, DEC, BayArea) und wir haben generalisierte phylogenetische Analysen der kleinsten Quadrate (PGLS) durchgeführt, um die möglichen Beziehungen zwischen Verbreitungsmustern, Nahrung, Körpermaßen, Gelegegröße und Zugverhalten festzustellen. Wir fanden, dass das wahrscheinlichste Herkunftsgebiet aller Turdus-Arten die östliche Paläarktis ist, gefolgt von einer sehr frühen Besiedlung der Westpaläarktis und von Afrika, und dass mehrere transatlantische Bewegungen zwischen 11-4 Millionen Jahren stattgefunden haben, also früher, als zuvor angenommen. Zugverhalten entstand schon in der Herkunftspopulation der Gattung Turdus, mit signifikanten Unterschieden in Gelegegröße und Nahrung von ziehenden und nicht-ziehenden Arten. Wir haben eine korrelierte Evolution zwischen Zugverhalten und der latitudinalen Ausbreitung, der hauptsächlichen Nahrung und saisonalem Nahrungswechsel. Wir schließen, dass sich die latitudinale Ausbreitung schon sehr früh entwickelt hat, etwa gleichzeitig mit der Radiation der Turdus-Drosseln, unmittelbar gefolgt von der Entwicklung meridionalen Zugverhaltens als Folge orographischer und klimatischer Veränderungen. Die bei Zugvogelarten beobachteten erhöhten Gelegegrößen und hier auch für die Drosseln, könnten wichtiger Kompensationsmechanismus zum Ausgleich der Mortalitätsverluste während des Zuges sein.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Recent research into the phylogenetic biogeography of the passerines (Fjeldså 2013; Payevsky 2014; Qu et al. 2014; Barker et al. 2015; Selvatti et al. 2015; Kennedy et al. 2016) has deepened our understanding of their distribution and the evolution of various life history traits. The estimation of ancestral areas, life history and behavioural traits based on the phylogeny of a species is a powerful approach (e.g. Nagy and Tökölyi 2014; Barker et al. 2015; Jonsson and Holt 2015; Selvatti et al. 2015; Moyle et al. 2016; Wang et al. 2016). Although thrushes (Turdidae) constitute an ideal group in which to explore such phenomena, relationships among lineages have long confounded taxonomists (e.g. Nylander et al. 2008; Voelker and Outlaw 2008; Olsson and Alström 2013). This suggests a need to review the phylogeny of this highly complex family of birds, and to carefully choose the taxa included in similar investigations.
More than 200 species have been classified into the Turdidae family (BirdLife International 2015a; see also the IOC World Bird List, Gill and Donsker 2018), half of which are considered to be Turdus or Zoothera, the two largest out of nearly 20 genera (Klicka et al. 2005). Although the migration and biogeography of several groups within the family have previously been studied (e.g. Nylander et al. 2008; Voelker et al. 2013), none of these studies used correlated evolutionary approaches to analyse the evolution of migration. The biogeographic assumptions also left underlying mechanisms to be clarified, especially in Turdus thrushes.
Phylogenetic relationships among thrush lineages and the dating of evolution within this group still remains uncertain, which also contrasts with reconstructions based on recently developed methods (e.g. Jetz et al. 2012). Accordingly, the beginning of Turdus radiation was dated to around 7 million years ago (Mya) by Nylander et al. (2008) and Voelker et al. (2009), while the more recent results of Jetz et al. (2012), based on all extant bird species, show that the earliest common ancestor of this group may have appeared at least 15 Mya. This contrasting date, up to twice the previous time interval, gives rise to new and so far unanswered questions regarding the biogeographic history of Turdus, especially when and how trans-Atlantic movements presumably occurred. Although hypotheses for possible paths have been formulated in the aforementioned works, these questions have not yet been fully answered, and a concise analysis and discussion of this topic are definitely needed.
The two biogeographically distinct groups of Zoothera were considered as polyphyletic taxa in Klicka et al. (2005), later corroborated by Jetz et al. (2012). Thus, a reconsideration of the biogeographic history of the whole Turdidae and the taxonomy of Zoothera thrushes is strongly recommended, as described below. Recently, all Geokichla have been classified as Zoothera; some species, e.g. Zoothera naevia and Zoothera pinicola have been categorized into this genus although they were previously considered the monotypic genera, Ixoreus naevius and Ridgwayia pinicola, respectively (Olsson and Alström 2013). According to the phylogenies of Jetz et al. (2012), Zoothera includes all former Geokichla species (e.g. Voelker and Outlaw 2008; Olsson and Alström 2013), split into two generic groups: the monophyletic Geokichla and the core Zoothera related to the subgenus Cochoa (see species list in Table S1). This latter group may also be considered as monophyletic assuming that the taxonomic position of the four, Catharus-related species, Zoothera naevia, Zoothera pinicola, Zoothera margaretae, and Zoothera leucolaema, will be revised later.
Considering these taxonomical uncertainties, it seems straightforward to only use Turdus (s.l.) as a model system including both Nesocichla eremita and Psophocichla litsitsirupa, and to recommend the above modifications for the attention of taxonomists. Furthermore, dissimilarities in the supposed timing of the evolution of Turdus require reconsideration of the evolutionary history of this genus by applying modern phylogenetic approaches based on molecular data (this study).
Moreover, little is known about the evolution of migration in thrushes (but see Winker and Pruett 2006; Voelker et al. 2013), which is hypothesized to be entangled with biogeographic events during the radiation of the passerines (see a general overview in Liedvogel and Delmore 2018). A number of migratory Turdus species have been recorded as regular visitors to areas outside of their normal distribution [e.g. Turdus iliacus, Turdus ruficollis, Turdus unicolor, but also the sedentary Turdus olivater, Turdus reeve, Turdus rufogalliatus (ABA Checklist version 7.9.0; Crochet and Joynt 2015)], and a very recent record is exemplified by Naumann’s Thrush (Turdus naumanni) in Hungary (Hadarics 2016), located at least 4500 km west of its known range. This phenomenon is common among passerines, e.g. also in buntings, warblers, and because these observations mainly represent along-latitudinal mobility, we hypothesize that migration along latitudes may be an earlier feature of these species (along-latitudinal first hypothesis), as an important trend in the speciation of this group. The correlation between genetic variation and migratory behaviour has also been reviewed by Pulido (2007). Furthermore, hypotheses on spatial segregation in birds have been previously discussed and spatial segregation found to be related to factors predicting migration strategy (e.g. Myers 1981; Smith et al. 2003; Mazerolle et al. 2005), body measurements (Belthoff and Gauthreaux 1991), and other factors (see a review on migratory systems by Faaborg et al. 2010; Liedvogel and Delmore 2018). Despite this, the diversification in birds suggested to have been promoted by the evolution of migration (Sol et al. 2005; Winger et al. 2012, 2014; Rolland et al. 2014; Fuchs et al. 2015).
Hence, examining the relationships between migration and factors of breeding phenology, body measurements, feeding ecology, and habitat selection may uncover novel patterns in the evolution and recent conditions of migration in Turdus thrushes. For example, the correlated evolution of migration with these components has never been studied. Feeding on different food sources is well known in birds, especially in small passerines, depending on the season (del Hoyo et al. 2016). These changes of feeding behaviour between seasons prompt us to ask the questions: what was the original behaviour of these birds, and how has it changed? Following previous results on changing foraging behaviour in sedentary/migratory lineages (Rainio et al. 2012; Telleria et al. 2013; Moyle et al. 2015) we formulated the insectivorous first hypothesis that predicts insectivory as an earlier state, predominantly in subtropical species, while the change from insectivory to herbivory in temperate zones implies transitions to migratory behaviour and vice versa. Similar to this, in those lineages where the ability for along-latitudinal movement occurs, stronger tendencies for migration are suspected for the reasons given in the previous paragraph.
To answer the questions given above, note that the evolutionary history of Turdus has been governed by major geological events that have had significant impacts on the global climate. Following the Paleocene–Eocene thermal maximum, climatic trends showed cooling to the lowest temperatures during the Oligocene with a smaller warm peak in the Middle Miocene (Zachos et al. 2001; Arnaiz-Villena et al. 2014). This interval coincides approximately with the appearance of the common ancestor of Turdus, and accelerating speciation in this group might be correlated with novel habitat types becoming available. The emergence of various savannah formations began during the Late Miocene in Africa (Bobe and Behrensmeyer 2004; Fuchs et al. 2015). The cooling climate and aridisation of North Africa led to the establishment of continental zonality of the continent in the Plio-Pleistocene, similar to present conditions (deMenocal 2004; Hernández Fernández and Vrba 2006), and provided an appropriate base for the colonization of new ecological niches. Trends of climatic cooling and glacial-interglacial phases over the past 15-10 million years (Zachos et al. 2001; Ravelo et al. 2004) may also have substantially driven biogeographic movements and the behavioural evolution of Turdus species, as seen in other groups [e.g. Carduelini (Arnaiz-Villena et al. 2014); Falconidae (Fuchs et al. 2015); Emberizidae (Päckert et al. 2015)].
The uplift of the Eastern Cordillera and Central Andes started around 10 Mya and continued until approximately 3 Mya (Gregory-Wodzicki 2000; Hooghiemstra and Van de Hammen 2004; Barke and Lamb 2006). These geological transformations and the climatic changes thereafter should have been sufficient to support the adaptive radiation of this group (Nylander et al. 2008; Voelker et al. 2009). The importance of studying this phenomenon has already been supported by other examples among the passerines: for instance, the basal branching in the Leptopogon genus occurred between 9 and 3 Mya (see Bates and Zink 1994 and discussion therein), while the uplift of the Andes also played a fundamental role in the divergence of the Thamnophilus genus (Brumfield and Edwards 2007).
The uplift of the Eastern Cordillera and the Andes could have enabled a pathway towards northern regions. However, the West Indies began to form in the Middle Eocene, and some of the major processes involved in this occurred after the Middle Miocene (Hedges 1996; Ricklefs and Bermingham 2008) when the present shape of these islands emerged. These events could form the basis of the colonization of the Caribbean Islands (e.g. Turdus plumbeus, Turdus jamaicensis, Turdus swalesi, Turdus lherminieri) and the dispersal into North America.
Thus, the aims of this study were: (1) to find an acceptable biogeographic explanation for the trans-Atlantic dispersal events of Turdus thrushes, (2) to examine their possible colonization paths in the Nearctic region in a revised time interval, and (3) to summarize the broad description of the worldwide biogeography of this group in the above-mentioned context (4) based on a comprehensive phylogeny of the genus. In the context of migration, we are also interested in (5) the evolutionary origin of migratory behaviour in Turdus thrushes (along-latitudinal first hypothesis); and (6) the relationships between life history traits, ecological factors (insectivorous first hypothesis), and migration within this group (e.g. Alerstam et al. 2003; Newton 2008).
Methods
Phylogenetic analyses
We mainly follow the descriptions in Nagy and Tökölyi (2014) and Johansson et al. (2018) with some modifications listed below. Gene sequences for 12S and 16S ribosomal RNA (12S, 16S, respectively), ATP synthase subunit 6 and subunit 8 (ATP6, ATP8, respectively), β-fibronigen intron 7 (BF7), cytochrome c oxidase subunit 1 (COX1), cytochrome b (CYTB), NADH-ubiquinone oxidoreductase subunit 2 and subunit 6 (ND2, ND6, respectively), and recombinase activating gene 1 (RAG1) were downloaded from GenBank (http://ncbi.nlm.nih.gov/). We searched data by the name of species following the taxonomic list of BirdLife International (2015a) due to a high matching rate of names in the database. However, the validity of taxa were also checked in IOC World Bird List (Gill and Donsker 2018). Available genes per species are summarised in Table S2. However, it is important to note that interpretation of phylogenies may differ depending on the data type used (Reddy et al. 2017; Nagy 2017), hence the support of some nodes in relation to the genetically under-represented species may be lower. Sequence alignment was done using MAFFT (Katoh et al. 2005) and cleaned with Gblocks (Castresana 2000). These steps were performed using ape (Paradis et al. 2004) and seqinr (Charif and Lobry 2007) packages in R version 3.2.4 (R Development Core Team 2016).
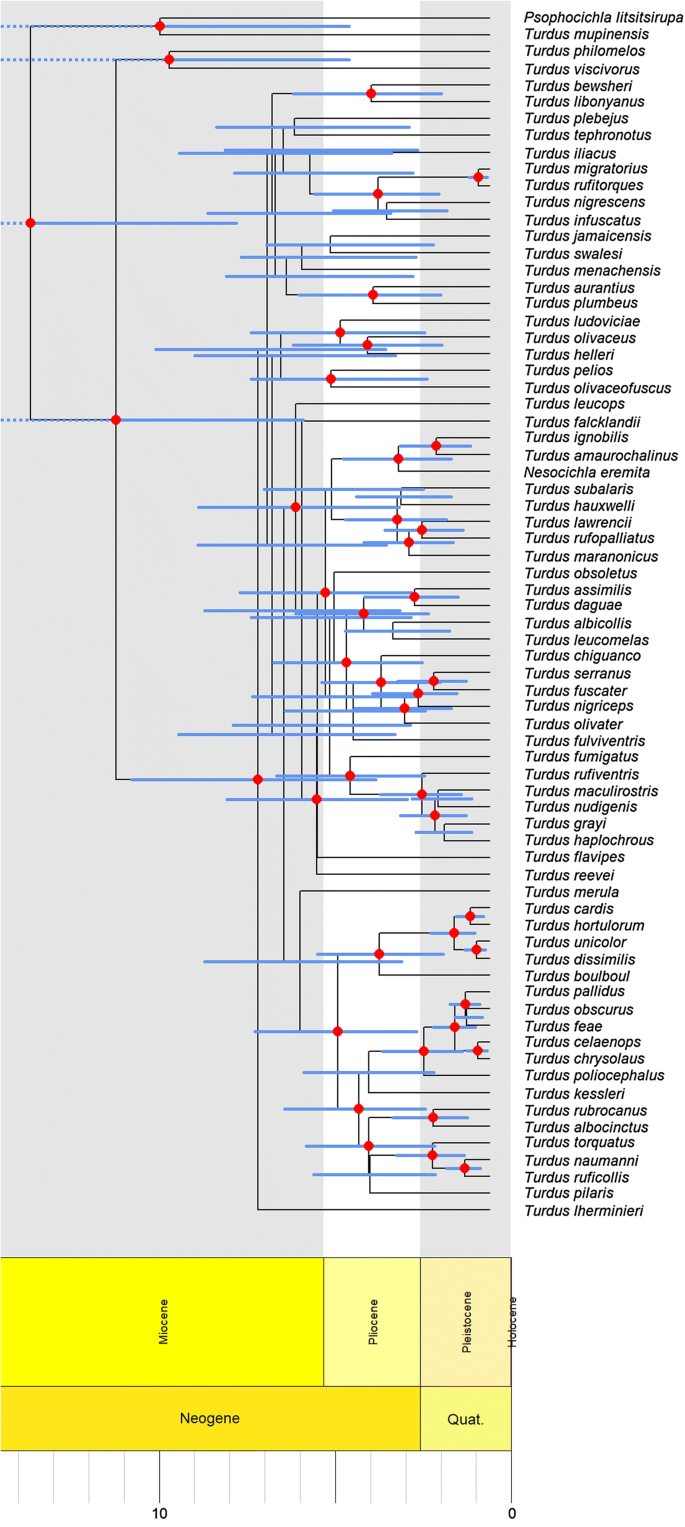
The randomised accelerated maximum likelihood approach (Stamatakis 2006) was applied to find the best scoring starting tree for the Bayesian Markov chain Monte Carlo analyses conducted in BEAST version 1.8.3 (Drummond and Rambaut 2007). The ten gene sequences were partitioned and the best-fitting substitution model according to the model test available in the phangorn package (Schliep 2011) was set for each (HKY+I–16S, RAG1; GTR–BF7; GTR+G—ND6; GTR+I–ATP6, ATP8; GTR+G+I—12S, COX1, CYTB, ND2). The uncorrelated relaxed molecular clocks approach was used for molecular dating (Drummond et al. 2006). Calibration priors based on fossil data were added following log normal distribution (mean = 0, SD = 0.5, with corresponding offsets), including all of the known oldest fossils among Turdus, to set divergent times to a minimum of 2.5 Mya for Turdus cf. rufiventris, Turdus cf. rufopalliatus, Turdus cf. ignobilis and Turdus cf. plebejus (Emslie 1998), 2.6 Mya for Turdus cf. pilaris, Turdus cf. philomelos and Turdus cf. torquatus [PaleoDB collection 124956, 124960 (Harrison 1987)], 3.5 Mya for Turdus cf. viscivorus and Turdus cf. merula (Clot et al. 1976), and 5.3 Mya for Turdus cf. iliacus (Jánossy 1991). In contrast to Nylander et al. (2008), we allowed the estimation of divergence times instead of setting them to a fixed point (6.8 Mya). However, Jánossy (1991) only considered an unspecific part of the Miocene and did not provide exact times nor intervals, thus we accounted for a minimum of 5.3 Mya for the age of the T. iliacus fossil, when the epoch approximately ended. These fossil records represent ancient occurrences of species in almost all recent biogeographic regions, except for the Nearctic where no data were found for records older than at least 1 million years. The BEAST run was allowed for 10,000,000 generations logging every 1000. The effective sample size of the parameters was checked in Tracer version 1.7.1. (Rambaut et al. 2018) and was higher than 200 for the majority of the parameters. However, in the case of a few parameters related to the substitution rates of under-represented genes (BF7, ND6), the values were closer to 100. Therefore, the initial settings applied for the phylogenetic analysis were considered to be acceptable. A maximum clade credibility tree (Fig. 1) was created from 10,000 trees after removing 10% burn-in using TreeAnnotator version 1.8.3. (Rambaut and Drummond 2016).
Maximum clade credibility tree of the studied 72 species. Estimated age ranges (95% highest posterior density intervals) are shown as blue bars and posterior probabilities > 0.8 are indicated by red dots at each node. The presented tree is rooted, ultrametric and all nodes are bifurcated (color figure online)
Biogeographic analyses
The availability of gene sequences and data on the ecology and life history of the species (listed below) limited the number of taxa included in our study, thus we collected breeding distribution data for 72 thrush species from BirdLife International (2015b) that comprehensively cover the whole Turdus genus out of the 200 species belonging to the Turdidae family (BirdLife International 2015a). Species were scored as present/absent in the following biogeographical realms/regions: Nearctic, Mexican, Neotropical, West and East Palearctic, Afrotropical, Indomalayan, Australasian, and Antarctic (Fig. S0). This regional classification forms operative biogeographic units for our analyses and should not be considered as identical to Wallace’s biogeographic subdivisions. Because many species are distributed in Mexico and one of the Nearctic or the Neotropical realms, we distinguished the Mexican region from the other large realms due to its significant position as a potential barrier/corridor. In our dataset, Turdus poliocephalus is the only species also distributed in some islands of the Australasian region, which suggests that Australasian species were under-represented among Turdus. However, it is clustered into an East Palearctic clade (see Figs. S0, S1–S5), thus its dispersal to these islands is more probable than the other way around. Therefore, we did not include Australasia as a separate biogeographic region (see also “Discussion”). Our own tree was used for the biogeographic analyses. We performed ancestral range estimation using probabilistic historical biogeography methods in the BioGeoBEARS package (Matzke 2013). The dispersal-vicariance (DIVA) (Ronquist 1997), dispersal-extinction-cladogenesis (DEC) (Ree et al. 2005; Ree and Smith 2008), and BayArea model (Landis et al. 2013), each complemented with founder-event speciation, were calculated and compared using the χ2-test. Biogeographical stochastic mapping (Matzke 2016) was accomplished and repeated 50 times for the model with the lowest Akaike information criterion value. The main differences among the three major biogeographic methods are indicated by the combinations of assumed processes: although all three models can basically deal with dispersal, extinction and narrow sympatry, in DIVA both narrow and widespread vicariance, in DEC subset sympatry and narrow vicariance, and in BayArea widespread sympatry, are also allowed to be estimated (Fig. S8).
We repeated the biogeographic analyses and compared the results using phylogenies for the subset of Turdus thrushes downloaded from Birdtree (http://birdtree.org/) where trees are generated using modified supertree and supermatrix approaches combining genetic and taxonomic information of all extant bird species (detailed description: http://birdtree.org/methods/). A consensus tree was generated using these trees (Jetz et al. 2012) that did not show lower posterior probabilities than either of the previously published ones (Klicka et al. 2005; Voelker et al. 2007, 2009; Nylander et al. 2008). Because the node ages showed high degrees of uncertainty based on the consensus tree from Birdtree.org, as well as on our tree, we repeated the biogeographic analyses for the majority rule consensus tree made by Nylander et al. (2008). However, Nylander et al. (2008) did not even provide detailed information of estimated time intervals, considering the facilitation of the containment of probable geological ages. The dating of previously published trees is supported by fossil records considered to be remains of thrushes (Jánossy 1991; Nylander et al. 2008) and fits the chronology of our phylogenetic estimation that was completed with the results of the biogeographic analyses below. This detailed comparison was particularly necessary to reveal consistencies in our ancestral biogeographic results based on phylogenies constructed with different methods. We conclude the same biogeographic history of the studied group independently from the phylogenies used (see Supplementary information, Additional information for the comparison of phylogenies).
Life history traits and migration
Data on migratory behaviour, body measurements, and life history-related traits were collected from several sources (see Table S3, del Hoyo et al. 2005; Clement and Hathway 2010; Rodewald 2016). The length of the breeding season was calculated using the start and end months for each species, while the difference between maximum and minimum altitude provided a range. Although thrushes are considered to be omnivores, diet breadth was calculated for both within and outside the breeding season as the sum of the consumed food types by species to measure differences in feeding behaviour (degree of omnivory). Change in food types between seasons was derived from the dissimilarity in main food types between the breeding season and outside this period. If the main food type of a species was the same in both seasons the score was 0 (which indicates no change), otherwise it was 1. Similar to this classification, when east–west (along-latitudinal) movement was described in the literature it was scored as 1 and in all other cases it was assigned 0. We considered this differentiation to be important because a significant proportion of the studied species (17%) could be assigned to the along-latitudinal movement category, which was either migratory or not.
We applied phylogenetic generalized least squares (PGLS) in the nlme package (Pinheiro et al. 2015) to analyse relationships between migratory behaviour and each paired explanatory variable. This design is especially important to find those single factors that are associated with migration and which could be included in the correlated evolutionary analyses described below. Two model sets were used as a basis for testing data fit. First, we ran all pairwise models for species (n = 60) for which data were available for all variables, and then we allowed the omission of missing values per variable (the number of included species varied between 66 and 72). The data structure was congruent in both sets, thus we ran models of species with data for all variables for 100 different phylogenetic trees. Best-fitted λ-values were generated from likelihood profiles of λ (Kamilar and Cooper 2013) derived from 500 runs for randomly selected values between 0 and 1, used dynamically in each PGLS run. A phylogenetic signal is a statistical measure of trait relatedness to phylogeny. Pagel’s λ (Pagel 1997, 1999) is considered a possible representation of a phylogenetic signal, hence λ = 0 indicates that phylogenetically closely related species are not more similar than distant relatives, whilst λ = 1 indicates a high phylogenetic signal (dependency) in the evolution of a given trait (Kamilar and Cooper 2013). We performed likelihood ratio tests to identify significant differences in models where λ was set to 0 and 1. Averaged λ, estimated value of parameters, SE, t- and p-values were calculated over the 100 runs, while the importance of significant and marginal variables was estimated by using model selection in the MuMIn package (Bartoń 2015). Those variables which proved to be significant after permitting variation of the sample size, and calculating the average values of 100 runs that allows one to control for the effect of phylogenetic history, should be considered as truly significant factors. All biogeographical and comparative analyses were performed in R version 3.2.4 (R Development Core Team 2016).
Evolution of migration
We investigated evolutionary changes and ancestral states in migratory behaviour. Non-migratory and migratory behaviour were classified as completely (all of the populations) and partially (some of the populations) migrant species within the latter category. First, we calculated transition rates between the categories coded as non-migratory and migratory. Second, ancestral state estimation was performed for the phylogeny using the maximum likelihood method to find the most probable state on the root, i.e. the ancient behaviour of the common ancestor of all Turdus. These analyses were done using the MultiState module of BayesTraits 2.0 (Pagel et al. 2004), which is applicable for estimating ancestral states and reconstructing how traits containing a few discrete states evolved on phylogenetic trees using a Bayesian approach and allowing maximum likelihood estimation of parameters.
We tested correlated evolution between migration and (1) main food type in, and out of, the breeding season; (2) shift in food type between seasons; and (3) the ability of the thrushes to move latitudinally. These analyses were run using the Discrete module of BayesTraits 2.0 (Pagel et al. 2004), a method which allows transition rates of correlated evolution to be estimated between a pair of traits in a phylogeny. These results complete the outcome of PGLS models regarding the aforementioned variables by which not only the estimation of correlation between traits is allowed but also the evaluation of well-defined directions that have significant information content for the evolution of migration. However, because this approach can only be applied to binary variables, we coded species as sedentary (0) and migratory (1). The same values were assigned to main food type (0—invertebrates, 1—fruits/seeds), to difference in consumed food type between seasons (0—the same type, 1—shift from insectivorous to herbivorous or vice versa), and to the ability to move latitudinally (0—not typical, 1—typical).
Furthermore, we mapped migratory behaviour on the phylogeny by applying stochastic character mapping for each trait simulated 1000 times in the phytools package (Revell 2012). All analyses were repeated on the partition of migratory behaviour from non-migratory, as well as partial and complete migration categories. As these two approaches are independent, BayesTraits and stochastic character mapping were expected to provide different results. The former is a robust method, while the latter approach allows better visualization tools for the estimated pattern.
Results
Biogeographic history of Turdus
Among the six biogeographical models, the DEC model with founder-event speciation fits the data the most parsimoniously (Table 1). All alternative models with founder-event speciation showed highly significant differences (p < 0.001) from the corresponding null model. According to the DEC+J model, the possible ancestral regions for all Turdus species are East Palearctic and/or Afrotropical realms, which appear in all of the acceptable combinations (see most probable state in Table 2). Two early diverging species (Turdus mupinensis, Psophocichla litsitsirupa), as well as the common ancestor of Turdus viscivorus and Turdus philomelos, suggest very early dispersal from Southeast Asia to Europe and Africa, or eventually vice versa. Nevertheless, the common ancestor of the remaining 68 species is assumed to be of Afrotropical/Neotropical origin, which suggests early, multiple trans-Atlantic movements approximately 10 and 9 Mya (nodes 77, 78, 92 and 96, Table 2). The ancestor of a large clade containing 19 species returned to Southeast Asia between 9.8 and 7 Mya. This event was followed by rapid speciation including the occupation of the Indomalayan region. Simultaneously, several repetitive trans-Atlantic movements are also supported by our analyses including both back and forth passage (main movements at nodes 82 and 115, respectively, Table 2; see also Fig. 2). These multiple events started around 11 and 9 Mya and continued until recent times, as the examples of Turdus lherminieri, Turdus plebejus, and Nesocichla eremita show (ages of the ancestors of these species are 11.02, 8.43 and 4.31 Mya, respectively). The youngest dispersal events relating to the colonization of Mexico and North America occurred in five independent lineages from South America (nodes 87, 90, 125, 133 and 139, Table 2) within the past 6–2 Mya.
Mapped phylogeny of Turdus thrushes indicating the major dispersal routes during their evolution. The most probable distribution of the common ancestor of Turdus mupinensis and Psophocichla litsitsirupa was in East Palearctic/Afrotropical regions (probability 0.4), whilst Turdus viscivorus and Turdus philomelos have ancient distributions in the East Palearctic region (probability 0.5)
The summary of 50 biogeographical stochastic mappings also supports the result of the DEC+J model (see Figs. S1, S2). Speciation events occurred 71 times during the evolution of Turdus thrushes (Fig. S3), separately for the possible processes as follows: anagenetic dispersal events (15.52 ± 1.23), sympatry (53.86 ± 1.92), subset sympatry (5.44 ± 2.12), founder event speciation (8.26 ± 1.43) and vicariance (3.44 ± 1.26).
Life history traits and migration
The results of our pairwise modelling show optimal data fits in all cases and similar significance values detected for the corresponding variables in both sets (Table S5–S6). Based on run values for 100 different phylogenies, we conclude that tree structure has no effect on estimations (see averaged values in Table S4). Estimation of λ does not show strong phylogenetic signal, nevertheless, the values range between 0.6 and 0.7 in many cases where the likelihood ratio test shows significant differences (< 0.001) from both of the models with pre-set λ-values, which implies that migratory behaviour and related traits probably evolved more dependently than independently among the lineages of Turdus thrushes.
Five out of the variables of substantial support were highly significant (p < 0.001). Considering the comparison of breeding ranges among species, migratory behaviour is more frequent in non-tropical species compared to those that breed in both climatic belts, and less frequent in tropical species than in species with breeding areas located in both tropical and non-tropical regions. Estimated parameters show that clutch size is higher in migratory species that exhibit shifts of main food type between breeding and non-breeding seasons. The important factors relevant to migration, based on the results of model selection, are also highly significant, which indicates that migratory species tend to be more mobile (represented by rarities), and most of them frequently move along a latitudinal (east–west) axis.
Biologically meaningful interactions among variables such as interactions among breeding range and factors of breeding phenology or habitat and dietary variables, had no significant effect on migratory behaviour. Furthermore, importance levels of variables estimated by model selection when migration was categorized into three classes were: 1 for breeding region, 0.99 for bill length, and 0.98 for the ability to move along latitudes. Repeating the analysis on migration in two categories showed the following importance levels: 1 for breeding belt, 0.91 for the ability to move along latitudes, 0.74 for bill length, and 0.65 for clutch size.
Evolution of migration
Out of the complete set of 72 thrush species, 55 species were classified as non-migratory, seven as partially migratory, and ten as complete migrants. Our results indicate that migration emerged as an ancestral behaviour in an early phase of Turdus evolution (Figs. S6, S7); the probability of migratory behaviour at the root of the phylogeny was estimated to be 0.52 in the analysis where this behaviour was split into two, and 0.67 for three categories (0.34 for partial and 0.33 for complete migration). Transition rates among states also confirm that migration was lost multiple times during evolution; the transition from non-migratory to one of the migratory states was estimated to be between 0 and 0.03, and the range from migratory states to a non-migratory state was 0.03–0.24. Furthermore, we found that in Turdus complete migration could only have evolved or been lost via partial migration (transition rates are 0.55 and 0.25, respectively).
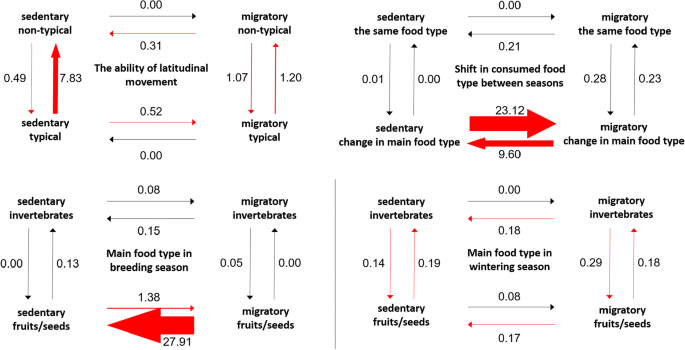
Correlated evolution between the ability to move along latitudes and migration shows that the most probable ancestral state of all species could be a combination of migratory and along-latitudinal movement (0.26), while neither of the two states of along-latitudinal movement were assigned similar probabilities (0.24). The appearance and disappearance of movements along latitudes thus could have frequently changed in migratory lineages (Fig. 3). The state of migratory behaviour is probably associated with the ability to move along latitudes, and the loss of this trait is most frequent in sedentary lineages.
Based on the analysis of migration and main food type utilized during the breeding season, transition rates show that sedentariness may have emerged more frequently than migratory behaviour in herbivorous lineages (Fig. 3). In contrast, a shift in the main food type from fruits/seeds to invertebrates is moderately typical of non-migratory lineages. Repeating this analysis based on the main food type between breeding seasons shows that the transition from fruits/seeds to invertebrates likely was at a higher rate, especially in migratory lineages, than vice versa (Fig. 3), the latter rate being almost equal to that of a transition between predominant food types from invertebrates to fruits/seeds in both lineages. The rates of changes in migratory behaviour in herbivorous lineages are likely to represent corresponding transitions in the main food type in the breeding season, however, with a substantially lower value. Based on data for the breeding season, the most probable ancestral state is a combination of sedentariness/migration and herbivory (both 0.32), but this cannot be clearly derived from the results of the analyses of data from outside the breeding season as probabilities for the combination of migratory/sedentary and insectivorous/herbivorous states are ca. 0.25 ± 0.01.
The analysis of migration and the shift in the main food type between seasons gives a similar estimation of ancestral states (i.e. the probability of all states is 0.25 ± 0.03). However, a change in the main food type is highly associated with changes in migratory behaviour, which indicates that migration could appear more frequently in lineages in which birds could have frequently changed food sources (Fig. 3). Furthermore, transition rates show a shift in diet from consumption of the same food type in both seasons to the frequent alternation between herbivorous and insectivorous behaviour in migratory lineages where inversion also has a similar rate.
Discussion
Our study provides new insights into (1) the biogeography of 72 Turdus thrushes, revealing the major biogeographic events behind the radiation of this group; (2) identifies the most probable paths in the colonization of newly available habitats; and (3) especially focuses on how the ability for movement along latitudes could have helped in this process and in the emergence of migration in this group. Furthermore, we discuss (4) correlated evolutionary scenarios between feeding and migration that allow us to explain several life history aspects including survival (the ability to find food under different climatic conditions and/or during migration), and (5) trade-offs between migration and reproductive success. In these respects, our study could be considered pioneering.
First, although global songbird radiation is considered to have originated in the Australasian region (e.g. Fjeldså 2013; Moyle et al. 2016; but see also Jonsson and Holt 2015), our findings provide new estimates of the biogeographical origin of the genus Turdus, by locating their most likely ancestral area in East Asia. According to our phylogenetic reconstruction, all basal nodes show posterior probabilities larger than 0.9, which suggests a more reliable biogeographic approach. Second, our results support our along-latitudinal first hypothesis for migration as a probable explanation for early colonization of the western Palearctic and Africa, followed by emerged and repeated trans-Atlantic events.
Specifically, we suggest that an along-latitudinal-first type of migratory strategy might have been possible in two different directions: either (1) along a northern route, through West Asia and Europe, which is supported by the distribution of the common ancestor of Turdus philomelos and Turdus viscivorus; or (2) via southward dispersal, through India, as supported by the ancient distribution of Turdus mupinensis and Psophocichla litsitsirupa. The latter two species form a sister group to all other Turdus, while the former are widely dispersed Palearctic species; these groups represent the two most ancient splits within the genus (Fig. 2). A high probability of mobility, inducing migration as an ancestral behavioural type within this group, is further confirmed by evidence of latitudinal dispersal, which possibly resulted in sympatric speciation (Mazerolle et al. 2005; Faaborg et al. 2010; but see also Winger et al. 2012, 2014). However, in the DEC model with founder event speciation, anagenetic events may be underestimated (Ree and Sanmartín 2018), but are not negligible in Turdus, according to the results of biogeographic stochastic mapping. This mode of speciation was also predicted for some Amazonian birds (e.g. Salisbury et al. 2012). The north–south migration could have emerged subsequently in thrushes as an adaptation to large-scale geological and climatic changes, which also led to an enhanced variation of climatic niche dynamics and a high diversification rate in some other groups (see e.g. Sol et al. 2005; Rolland et al. 2014; Fuchs et al. 2015; Cooney et al. 2016; Gómez et al. 2016). To further support the above explanation, examples can be seen among rarities and in the distribution patterns of many migratory species. Birds observed as rarities in North America mainly originated from Europe, and rarities in Europe are usually western marginal occurrences of East Asian species (Crochet and Joynt 2015; ABA 2016). Furthermore, the density of migratory Turdus species (darker colours, Fig. S9) shows that many species breed in an extended area along an east–west axis in the Palearctic region and winter far west of their easternmost breeding distribution (BirdLife International 2015b).
In contrast to the above, orogenesis prior to the Late Tertiary, in combination with the uplift of the Tibetan (Qinghai-Tibet) Plateau, may have forced the lineage to move along an east–west axis in Asia. Arnaiz-Villena et al. (2014) suggested that changes during the Miocene may have had a significant impact on the evolution of Carduelini finches, and this geological period may have also been influential in accentors (Prunellidae) (Drovetski et al. 2013), in a radiation that might parallel the evolutionary history of Turdus [additional examples can be seen in warblers (Parulidae and Phylloscopidae) (Price 2010; Winger et al. 2012; Bloch et al. 2015), and buntings (Emberizidae) Päckert et al. 2015]. Orogenic events in the Late Tertiary [especially the uplift of the Andes (Gregory-Wodzicki 2000; Hooghiemstra and Van de Hammen 2004; Barke and Lamb 2006)] formed possible routes for South American biota to access the north. Global cooling in the Upper Miocene (Zachos et al. 2001) drove these species to develop migratory behaviour as a consequence of climatic constraints in the Northern Hemisphere. Movements along latitudes thus could have evolved earlier, facilitated by the more contiguous tropical zone from East Asia through northern Africa to South America, providing a corridor for the radiation of Turdus thrushes, followed by the appearance of meridional migration within the group as increased genetic differences due to speciation, which may have been a response to variation in migratory behaviour (Sol et al. 2005; Pulido 2007; Winger et al. 2012, 2014). A significant correlation between migratory behaviour and speciation rate has also been detected in other bird groups [e.g. some groups of New World songbirds (Winger et al. 2014); harriers (Oatley et al. 2015); falconid raptors (Fuchs et al. 2015)], but also in Catharus, considered a sister group of Turdus (Outlaw et al. 2003; Winker and Pruett 2006).
Based on the results and the chronology of phylogenetic trees used in our study, dispersal to South America may have occurred from the direction of Africa rather than from North America, which is in accordance with the South American origin of White-throated Thrush (Turdus assimilis) and its allies (Núñez-Zapata et al. 2016). The larger set of Neotropical species form a younger clade in our analyses with possibly Afrotropical ancestors, thus northern colonization routes are unlikely (Fig. S0). However, in the case of Turdus iliacus, a species nested in the older Neotropical clade, together with some North American and other Eurasian species, a probable trans-Beringian resettlement of Eurasian regions is possible, the pattern of which can be found in, e.g. Winter Wren (Troglodytes troglodytes), Pine Grosbeak (Pinicola enucleator), among passerines (Drovetski et al. 2004, 2010), and also in Three-toed Woodpecker (Picoides tridactylus) (Zink et al. 1995, 2002). Furthermore, Catharus thrushes, an outgroup of all species in our dataset (see Table S1 and also Outlaw et al. 2003; Winker and Pruett 2006), could have originated from North America (Voelker et al. 2013) after the split from Turdus, and colonization of the northern regions from the direction of Asia. Nevertheless, a Central or South American origin is supported for the Catharus (Outlaw et al. 2003), which is in line with our results. Moreover, the highest uncertainties were found in the large clade containing mainly Neotropical and Afrotropical species, which may be due to either the under-representation of genetic information for these species or the frequent mixing between the populations on both sides of the Atlantic Ocean that resulted in infering histories of the studied gene sequences (Reddy et al. 2017; Nagy 2017). Therefore, these contradictory patterns call for the re-analysis of the biogeographic history of the Turdidae after a deep taxonomic revision, as suggested in the Introduction.
Previous studies have shown that species are able to remain migratory if they occupy boreal regions, including mountainous habitats. For example, in the Carduelini tribe, most species are distributed in either mountainous or boreal areas (Arnaiz-Villena et al. 2001; BirdLife International 2015b), while migratory/highly mobile species frequently move on an axis along the latitudes (also Phylloscopus sp.). However, the loss of migration in crossbills (Loxia spp.) compared to their sister taxon, including migratory Carduelis species, may have been driven by their adaptation to feeding on the seeds of pine cones (Arnaiz-Villena et al. 2001; Benkman 2003; Parchman et al. 2006; Nguembock et al. 2009; Thompson 2016). The Siberian accentor (Prunella montanella), a long-distance migrant between Siberia and the Tibetan Plateau and also a regular visitor to Western Europe and North America, is a member of an avian family that includes many non-migrant species (Drovetski et al. 2013; del Hoyo et al. 2016). A risk-avoidance strategy is more likely in similar cases where migratory species move on latitudinal axis rather than crossing the Himalayas [e.g. Prunella and Phylloscopus (Price 2010; Drovetski et al. 2013)]. The loss of migration thus seems to be more frequent during the evolution of similarly behaving passerine groups, as the loss of migratory behaviour is often related to changes in foraging (e.g. Moyle et al. 2015; but see also Fuchs et al. 2015). Correlated evolution between migration and food types, distinguishing seasonal shifts in the latter, predicts important plasticity in response to the changing climate of geological history. In Turdus thrushes, shifts from consuming the same food type or switching food type between breeding and wintering seasons were relatively common in migratory lineages, and could have been the result of flexibility to changing environmental conditions during migration and on breeding/wintering sites. Species with the ability to easily switch food sources could change their migratory behaviour more frequently [see e.g. in the Parulidae (Gómez et al. 2016), or the synthesis by Cooney et al. (2016)]. Although sedentary Turdus species have mainly tropical/subtropical distributions (BirdLife International 2015b; del Hoyo et al. 2016), the insectivorous first hypothesis has been proposed as a reasonable explanation for the sedentariness of herbivorous species, including frugivorous taxa, due to changes in the main food type that occurred frequently in migratory lineages (Rainio et al. 2012; Telleria et al. 2013; but see Moyle et al. 2015). However, note that we are unable to clearly define complete causal relationships, and are only able to highlight possible changes between evolutionary states; further investigation is required to further explore this correlation.
Further evidence supporting a shift in main food type between seasons in passerine species has been presented in this analysis (similarly to e.g. Rainio et al. 2012; Telleria et al. 2013). Correlated evolutionary analyses showed that, among migratory thrushes, shifts from insectivorous to frugivorous behaviour during the non-breeding season might have been frequent during evolution, the flexibility of which is supported by many examples (e.g. Carpodacus erythrinus, Emberiza koslowi, Prunella himalayana) of feeding behaviour in present bird species (del Hoyo et al. 2016). A clear trend in a recent shift in food types emerges not only in resident species but also in migratory birds, as they are increasingly dependent on plant matter, including fruits (Telleria et al. 2013). Further work on bill shape and size, which should be highly correlated with consumed food type, may support these findings.
In addition, whether a bird is a herbivorous resident or an insectivorous migrant has important consequences for its conservation (Rainio et al. 2012). It has long been known that there are important trade-offs between migration and survival (especially reproductive success) (Alerstam et al. 2003; Newton 2008); for example, in a case study of Wood Thrushes (Hylocichla mustelina), resident birds had higher survival rates than vagrants (Rappole et al. 1989), as migrants are more frequently exposed to mortality factors than residents (e.g. Ketterson and Nolan 1982; Bell 1996; Alerstam et al. 2003; Newton 2008). Strategies related to migration have already been suggested as one of the possible explanations for the variation in adult survival among Turdus and Catharus species (Boyce and Martin 2017), e.g. the increased clutch sizes seen in migratory species, as documented here for Turdus (for other examples, see Martin 1995; Jahn and Cueto 2012; Barve and Mason 2015; Nagy et al. 2017), may be a mechanism to offset mortality during migration.
In conclusion, using integrative data on the distribution and life history of Turdus thrushes, we obtained (1) robust results for the biogeographical origin of the genus that support our along-latitudinal first hypothesis for migration as a probable explanation for this group’s distribution pattern in the early colonization of the western Palearctic and South America through Africa, and (2) found correlations between migration and food types that may have been important for the evolution of these birds.
References
ABA (2016) The ABA checklist version 7.9.0. American Birding Association, Delaware City. http://listing.aba.org/aba-checklist/. Accessed 17 Aug 2016
Alerstam T, Hedenström A, Akesson S (2003) Long-distance migration: evolution and determinants. Oikos 103:247–260
Arnaiz-Villena A, Guillén J, Ruiz-del-Valle V, Lowy E, Zamora J, Varela P, Stefani D, Allende LM (2001) Phylogeography of crossbills, bullfinches, grosbeaks, and rosefinches. CMLS Cell Mol Life Sci 58:1–8
Arnaiz-Villena A, Ruiz-del-Valle V, Gomez-Prieto P, Rey D, Enriquez-de-Salamanca M, Marco J, Muniz E, Martin-Villa M, Areces C (2014) Carduelini new sistematics: Crimson-winged Finch (Rhodopechys sanguineus) is included in “arid-zone” Carduelini finches by mitochondrial DNA phylogeny. Open Ornithol J 7:55–62
Barke R, Lamb S (2006) Late Cenozoic uplift of the Eastern Cordillera, Bolivian Andes. Earth Planet Sci Lett 249:350–367
Barker FK, Burns KJ, Klicka J, Lanyon SM, Lovette IJ (2015) New insights into New World biogeography: an integrated view from the phylogeny of blackbirds, cardinals, sparrows, tanagers, warblers, and allies. Auk 132:333–348
Bartoń K (2015) MuMIn: multi-model inference. R package version 1.13.4. https://cran.r-project.org/web/packages/. Accessed 23 Nov 2016
Barve S, Mason NA (2015) Interspecific competition affects evolutionary links between cavity nesting, migration and clutch size in Old World flycatchers (Muscicapdae). Ibis 157:299–311
Bates JM, Zink RM (1994) Evolution into the Andes: molecular evidence for species relationships in the genus Leptopogon. Auk 111:507–515
Bell CP (1996) The relationship between geographic variation in clutch size and migration pattern in the Yellow Wagtail. Bird Study 43:333–341
Belthoff JR, Gauthreaux SA (1991) Partial migration and differential winter distribution of House Finches in the eastern United States. Condor 93:374–382
Benkman CW (2003) Divergent selection drives the adaptive radiation of crossbills. Evolution 57:1176–1181
BirdLife International (2015a) The BirdLife checklist of the birds of the world: version 8. http://datazone.birdlife.org/home. Accessed 29 Oct 2015
BirdLife International (2015b) IUCN Red List for birds. http://datazone.birdlife.org/home. Accessed 29 Oct 2015
Bloch NI, Morrow JM, Chang BSW, Price TD (2015) SWS2 visual pigment evolution as a test of historically contingent patterns of plumage color evolution in warblers. Evolution 69:341–356
Bobe R, Behrensmeyer AK (2004) The expansion of grassland ecosystems in Africa in relation to mammalian evolution and the origin of the genus Homo. Palaeogeogr Palaeoclimatol Palaeoecol 207:399–420
Boyce AJ, Martin TE (2017) Contrasting latitudinal patterns of life history divergence in two genera of New World thrushes (Turdinae). J Avian Biol 48:581–590
Brumfield RT, Edwards SV (2007) Evolution into and out of the Andes: a Bayesian analysis of historical diversification in Thamnophilus antshrikes. Evolution 61:346–367
Castresana J (2000) Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol 17:540–552
Charif D, Lobry JR (2007) SeqinR 1.0-2: a contributed package to the R project for statistical computing devoted to biological sequences retrieval and analysis. Structural approaches to sequence evolution. Springer, Berlin, Heidelberg, pp 207–232
Clement P, Hathway R (2010) Thrushes. Bloomsbury, London
Clot A, Chaline J, Heintz E, Jammot D, Mourer-Chauviré C, Rage JC (1976) Montoussé 5 (Hautes-Pyrénées), un nouveau remplissage de fissure à faune de vertébrés du Pléistocène inférieur. Geobios 9:511–514
Cooney CR, Seddon N, Tobias JA (2016) Widespread correlations between climatic niche evolution and species diversification in birds. J Anim Ecol 85:869–878
Crochet PA, Joynt G (2015) AERC list of Western Palearctic birds. http://www.aerc.eu/tac.html. Accessed 19 Aug 2016
del Hoyo J, Elliot A, Christie DA (2005) Handbook of the birds of the world (Volume 10): cuckoo-shrikes to thrushes. Lynx, Barcelona
del Hoyo J, Elliott A, Sargatal J, Christie DA, de Juana E (2016) Handbook of the birds of the world alive. Lynx, Barcelona
deMenocal PB (2004) African climate change and faunal evolution during the Pliocene–Pleistocene. Earth Planet Sci Lett 220:3–24
Drovetski SV, Zink RM, Rohwer S, Fadeev IV, Nesterov EV, Karagodin I, Koblik EA, Red’kin YA (2004) Complex biogeographic history of a Holarctic passerine. Proc R Soc Lond B Biol Sci 271:545–551
Drovetski SV, Zink RM, Ericson PGP, Fadeev IV (2010) A multilocus study of Pine Grosbeak phylogeography supports the pattern of greater intercontinental divergence in Holarctic boreal forest birds than in birds inhabiting other high-latitude habitats. J Biogeogr 37:696–706
Drovetski SV, Semenov G, Drovetskaya SS, Fadeev IV, Red’kin YA YA, Voelker G (2013) Geographic mode of speciation in a mountain specialist avian family endemic to the Palearctic. Ecol Evol 3:1518–1528
Drummond AJ, Rambaut A (2007) BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol 7:214
Drummond AJ, Ho SY, Phillips MJ, Rambaut A (2006) Relaxed phylogenetics and dating with confidence. PLoS Biol 4:e88
Emslie SD (1998) Avian community, climate, and sea-level changes in the Plio-Pleistocene of the Florida Peninsula. Ornithol Monogr 50:1–113
Faaborg J, Holmes RT, Anders AD, Bildstein KL, Dugger KM, Gauthreaux SA, Heglund P, Hobson KA, Jahn AE, Johnson DH, Latta SC, Levey DJ, Marra PP, Merkord CL, Nol E, Rothstein SI, Sherry TW, Sillett TS, Thompson FR, Warnock N (2010) Recent advances in understanding migration systems of New World land birds. Ecol Monogr 80:3–48
Fjeldså J (2013) The global diversification of songbirds (Oscines) and the build-up of the Sino-Himalayan diversity hotspot. Chin Birds 4:132–143
Fuchs J, Johnson JA, Mindell DP (2015) Rapid diversification of falcons (Aves: Falconidae) due to expansion of open habitats in the Late Miocene. Mol Phylogenet Evol 82:166–182
Gill F, Donsker D (2018) IOC world bird list (version 8.2). https://doi.org/10.14344/IOC.ML.8.2; https://www.worldbirdnames.org/
Gómez C, Tenorio EA, Montoya P, Cadena CD (2016) Niche-tracking migrants and niche-switching residents: evolution of climatic niches in New World warblers (Parulidae). Proc R Soc Lond B Biol Sci 283:20152458
Gregory-Wodzicki KM (2000) Uplift history of the Central and Northern Andes: a review. GSA Bull 112:1097–1105
Hadarics T (2016) New species in the Hungarian avifauna in 2015. Ornis Hung 24:78–83
Harrison CJO (1987) Pleistocene and prehistoric birds of south-west Britain. Proc Univ Bristol Spelaeol Soc 18:81–104
Hedges SB (1996) Historical biogeography of West Indian vertebrates. Annu Rev Ecol Syst 27:163–196
Hernández Fernández M, Vrba ES (2006) Plio-Pleistocene climatic change in the Turkana Basin (East Africa): evidence from large mammal faunas. J Hum Evol 50:595–626
Hooghiemstra H, Van de Hammen T (2004) Quaternary Ice-Age dynamics in the Colombian Andes: developing an understanding of our legacy. Philos Trans R Soc Lond B 359:173–181
Jahn AE, Cueto VR (2012) The potential for comparative research across New World bird migration systems. J Ornithol 153:199–205
Jánossy D (1991) Late Miocene bird remains from Polgárdi (W-Hungary). Aquila 98:13–35
Jetz W, Thomas GH, Joy JB, Hartmann K, Mooers AO (2012) The global diversity of birds in space and time. Nature 491:444–448
Johansson US, Nylinder S, Ohlson JI, Tietze DT (2018) Reconstruction of the late Miocene biogeographical history of tits and chickadees (Aves: Passeriformes: Paridae): a comparison between discrete area analyses and probabilistic diffusion approach. J Biogeogr 45:14–25
Jonsson KA, Holt BG (2015) Islands contribute disproportionately high amounts of evolutionary diversity in passerine birds. Nat Commun 6:8538
Kamilar JM, Cooper N (2013) Phylogenetic signal in primate behaviour, ecology and life history. Philos Trans R Soc B 368:20120341
Katoh K, Kuma KI, Toh H, Miyata T (2005) MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res 33:511–518
Kennedy JD, Price TD, Fjeldså J, Rahbek C (2016) Historical limits on species co-occurrence determine variation in clade richness among New World passerine birds. J Biogeogr 44:736–747
Ketterson ED, Nolan V (1982) The role of migration and winter mortality in the life history of a temperate-zone migrant, the Dark-eyed Junco, as determined from demographic analyses of winter populations. Auk 99:243–259
Klicka J, Voelker G, Spellman GM (2005) A molecular phylogenetic analysis of the “true thrushes” (Aves: Turdinae). Mol Phylogenet Evol 34:486–500
Landis MJ, Matzke NJ, Moore BR, Huelsenbeck JP (2013) Bayesian analysis of biogeography when the number of areas is large. Syst Biol 62:789–804
Liedvogel M, Delmore K (2018) (Micro) evolutionary changes and the evolutionary potential of bird migration. In: Tietze DT (ed) Bird species. Springer, Cham, pp 109–127
Martin TE (1995) Avian life history evolution in relation to nest sites, nest predation, and food. Ecol Monogr 65:101–127
Matzke NJ (2013) Probabilistic historical biogeography: new models for founder-event speciation, imperfect detection, and fossils allow improved accuracy and model-testing. Ph.D. thesis, Department of Integrative Biology and Designated Emphasis in Computational and Genomic Biology, University of California, Berkeley
Matzke NJ (2016) Stochastic mapping under biogeographical models. PhyloWiki BioGeoBEARS. http://phylo.wikidot.com/. Accessed 15 Jan 2018
Mazerolle DF, Hobson KA, Wassenaar LI (2005) Stable isotope and band-encounter analyses delineate migratory patterns and catchment areas of White-throated Sparrows at a migration monitoring station. Oecologia 144:541–549
Moyle RG, Hosner PA, Jones AW, Outlaw DC (2015) Phylogeny and biogeography of Ficedula flycatchers (Aves: Muscicapidae): novel results from fresh source material. Mol Phylogenet Evol 82:87–94
Moyle RG, Oliveros CH, Andersen MJ, Hosner PA, Benz BW, Manthey JD, Travers SL, Brown RM, Faircloth BC (2016) Tectonic collision and uplift of Wallacea triggered the global songbird radiation. Nat Commun 7:12790
Myers JP (1981) A test of three hypotheses for latitudinal segregation of the sexes in wintering birds. Can J Zool 59:1527–1534
Nagy J (2017) Phylogeny and evolution of the European Goldfinch (Carduelis carduelis) and its allies–a review of the “bird of the year”. Ornis Hung 25:1–10
Nagy J, Tökölyi J (2014) Phylogeny, historical biogeography and the evolution of migration in accipitrid birds of prey (Aves: Accipitriformes). Ornis Hung 22:15–35
Nagy J, Zs Végvári, Varga Z (2017) Life history traits, bio-climate and migratory systems of accipitrid birds of prey (Aves: Accipitriformes). Biol J Linn Soc 121:63–71
Newton I (2008) The migration ecology of birds. Academic Press, London
Nguembock B, Fjeldså J, Couloux A, Pasquet E (2009) Molecular phylogeny of Carduelinae (Aves, Passeriformes, Fringillidae) proves polyphyletic origin of the genera Serinus and Carduelis and suggests redefined generic limits. Mol Phylogenet Evol 51:169–181
Núñez-Zapata J, Peterson AT, Navarro-Sigüenza AG (2016) Pleistocene diversification and speciation of White-throated Thrush (Turdus assimilis; Aves: Turdidae). J Ornithol 157:1073–1085
Nylander JAA, Olsson U, Alström P, Sanmartin I (2008) Accounting for phylogenetic uncertainty in biogeography: a Bayesian approach to dispersal-vicariance analysis of the thrushes (Aves: Turdus). Syst Biol 57:257–268
Oatley G, Simmons RE, Fuchs J (2015) A molecular phylogeny of the harriers (Circus, Accipitridae) indicate the role of long-distance dispersal and migration in diversification. Mol Phylogenet Evol 85:150–160
Olsson U, Alström P (2013) Molecular evidence suggests that the enigmatic Sulawesi endemic Geomalia heinrichi belongs in the genus Zoothera (Turdidae, Aves). Chin Birds 4:155–160
Outlaw DC, Voelker G, Mila B, Girman DJ (2003) Evolution of long-distance migration in and historical biogeography of Catharus thrushes: a molecular phylogenetic approach. Auk 120:299–310
Päckert M, Sun Y, Strutzenberger P, Valchuk O, Tietze DT, Martens J (2015) Phylogenetic relationships of endemic bunting species (Aves, Passeriformes, Emberizidae, Emberiza koslowi) from the eastern Qinghai-Tibet Plateau. Vertebr Zool 65:135–150
Pagel M (1997) Inferring evolutionary processes from phylogenies. Zool Scr 26:331–348
Pagel M (1999) Inferring the historical patterns of biological evolution. Nature 401:877
Pagel M, Meade A, Barker D (2004) Bayesian estimation of ancestral character states on phylogenies. Syst Biol 53:673–684
Paradis E, Claude J, Strimmer K (2004) APE: analyses of phylogenetics and evolution in R language. Bioinformatics 20:289–290
Parchman TL, Benkman CW, Britch SC (2006) Patterns of genetic variation in the adaptive radiation of New World crossbills (Aves: Loxia). Mol Ecol 15:1873–1887
Payevsky VA (2014) Phylogeny and classification of passerine birds, Passeriformes. Biol Bull Rev 4:143–156
Pinheiro J, Bates D, DebRoy S, Sarkar D, R Development Core Team (2015) nlme: linear and nonlinear mixed effects models. R package version 3.1-121. https://cran.r-project.org/web/packages/. Accessed 13 Jan 2018
Price TD (2010) The roles of time and ecology in the continental radiation of the Old World leaf warblers (Phylloscopus and Seicercus). Philos Trans R Soc Lond B 365:1749–1762
Pulido F (2007) The genetics and evolution of avian migration. AIBS Bull 57:165–174
Qu Y, Song G, Gao B, Quan Q, Ericson PGP, Lei F (2014) The influence of geological events on the endemism of East Asian birds studied through comparative phylogeography. J Biogeogr 42:179–192
R Development Core Team (2016) R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna. https://www.r-project.org/. Accessed 13 Mar 2016
Rainio MJ, Kanerva M, Wahlberg N, Nikinmaa M, Eeva T (2012) Variation of basal EROD activities in ten passerine bird species—relationships with diet and migration status. PLoS One 7:e33926
Rambaut A, Drummond AJ (2016) TreeAnnotator version 1.8.3. University of Edinburgh, Edinburgh. http://beast.community/programs. Accessed 4 Nov 2016
Rambaut A, Drummond AJ, Xie D, Baele G, Suchard MA (2018) Posterior summarisation in Bayesian phylogenetics using Tracer 1.7. Syst Biol 67:901–904
Rappole JH, Ramos MA, Winker K (1989) Wintering Wood Thrush movements and mortality in Southern Veracruz. Auk 106:402–410
Ravelo AC, Andreasen DK, Lyle M, Lyle AO, Wara MW (2004) Regional climate shifts caused by gradual global cooling in the Pliocene epoch. Nature 429:263–267
Reddy S, Kimball RT, Pandey A, Hosner PA, Braun MJ, Hackett SJ, Han K, Harshman J, Huddleston CJ, Kingston S, Marks BD, Miglia KJ, Moore WS, Sheldon FH, Witt CC, Yuri T, Braun EL (2017) Why do phylogenomic data sets yield conflicting trees? Data type influences the avian tree of life more than taxon sampling. Syst Biol 66:857–879
Ree RH, Sanmartín I (2018) Conceptual and statistical problems with the DEC+J model of founder-event speciation and its comparison with DEC via model selection. J Biogeogr 45:741–749
Ree RH, Smith SA (2008) Maximum likelihood inference of geographic range evolution by dispersal, local extinction, and cladogenesis. Syst Biol 57:4–14
Ree RH, Moore BR, Webb CO, Donoghue MJ (2005) A likelihood framework for inferring the evolution of geographic range on phylogenetic trees. Evolution 59:2299–2311
Revell LJ (2012) phytools: an R package for phylogenetic comparative biology (and other things). Methods Ecol Evol 3:217–223
Ricklefs R, Bermingham E (2008) The West Indies as a laboratory of biogeography and evolution. Philos Trans R Soc Lond B 363:2393–2413
Rodewald P (2016) The birds of North America online. Cornell Laboratory of Ornithology, Ithaca
Rolland J, Jiguet F, Jonsson KA, Condamine FL, Morlon H (2014) Settling down of seasonal migrants promotes bird diversification. Proc R Soc B 281:20140473
Ronquist F (1997) Dispersal-vicariance analysis: a new approach to the quantification of historical biogeography. Syst Biol 46:195–203
Salisbury CL, Seddon N, Cooney CR, Tobias JA (2012) The latitudinal gradient in dispersal constraints: ecological specialisation drives diversification in tropical birds. Ecol Lett 15:847–855
Schliep KP (2011) phangorn: phylogenetic analysis in R. Bioinformatics 27:592–593
Selvatti AP, Gonzaga LP, de Moraes Russo CA (2015) A Paleogene origin for crown passerines and the diversification of the Oscines in the New World. Mol Phylogenet Evol 88:1–15
Smith RB, Meehan TD, Wolf BO (2003) Assessing migration patterns of Sharp-shinned Hawks Accipiter striatus using stable-isotope and band encounter analysis. J Avian Biol 34:387–392
Sol D, Stirling DG, Lefebvre L (2005) Behavioural drive or behavioural inhibition in evolution: subspecific diversification in Holarctic passerines. Evolution 59:2669–2677
Stamatakis A (2006) RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22:2688–2690
Telleria JL, Blázquez M, De La Hera I, Pérez-Tris J (2013) Migratory and resident Blackcaps Sylvia atricapilla wintering in southern Spain show no resource partitioning. Ibis 155:750–761
Thompson JN (2016) Coevolution, local adaptation and ecological speciation. Mol Ecol 25:5608–5610
Voelker G, Outlaw RK (2008) Establishing a perimeter position: speciation around the Indian Ocean Basin. J Evol Biol 21:1779–1788
Voelker G, Rohwer S, Bowie RCK, Outlaw DC (2007) Molecular systematics of a speciose, cosmopolitan songbird genus: defining the limits of, and relationships among, the Turdus thrushes. Mol Phylogenet Evol 42:422–434
Voelker G, Rohwer S, Outlaw DC, Bowie RCK (2009) Repeated trans-Atlantic dispersal catalysed a global songbird radiation. Glob Ecol Biogeogr 18:41–49
Voelker G, Bowie RCK, Klicka J (2013) Gene trees, species trees and Earth history combine to shed light on the evolution of migration in a model avian system. Mol Ecol 22:3333–3344
Wang N, Kimball RT, Braun EL, Liang B, Zhang Z (2016) Ancestral range reconstruction of Galliformes: the effects of topology and taxon sampling. J Biogeogr 44:122–135
Winger BM, Lovette IJ, Winker DW (2012) Ancestry and evolution of seasonal migration in the Parulidae. Proc R Soc B 279:610–618
Winger BM, Barker FK, Ree RH (2014) Temperate origins of long-distance seasonal migration in New World songbirds. Proc Natl Acad Sci 111:12115–12120
Winker K, Pruett CL (2006) Seasonal migration, speciation, and morphological convergence in the genus Catharus (Turdidae). Auk 123:1052–1068
Zachos J, Pagani M, Sloan S, Thomas E, Billups K (2001) Trends, rhythms, and aberrations in global climate 65 Ma to present. Science 292:686–693
Zink RM, Rohwer S, Andreev AV, Dittmann DL (1995) Trans-Beringia comparisons of mitochondrial DNA differentiation in birds. Condor 97:639–649
Zink RM, Rohwer S, Drovetski S, Blackwell-Rago RC, Farrell SL (2002) Holarctic phylogeography and species limits of Three-toed Woodpeckers. Condor 104:167–170
Acknowledgements
We are grateful to our friends and colleagues for their suggestions on an earlier version of this paper. Many thanks go to Mark C. Mainwaring and to Adam Benlemlih for language revision, to Johan A. A. Nylander for providing access to his published phylogeny, and to our colleagues in the András Keve Library for Ornithology and Nature Conservation for allowing access to their printed literature. We also thank the two anonymous reviewers for their useful comments and suggestions. Open access funding provided by University of Debrecen (DE).
Author information
Authors and Affiliations
Contributions
J. N. collected and analysed the data, designed the methods and conducted the research. All of the authors took part in the formation of questions and hypotheses. J. N. wrote the paper in cooperation with Z. V. and Z. V.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflicts of interest.
Ethics statement
All data and information from external sources used for the analyses and preparation of the manuscript are properly cited. The conclusions are entirely based on our own results.
Additional information
Communicated by M. Wink.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Nagy, J., Végvári, Z. & Varga, Z. Phylogeny, migration and life history: filling the gaps in the origin and biogeography of the Turdus thrushes. J Ornithol 160, 529–543 (2019). https://doi.org/10.1007/s10336-019-01632-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10336-019-01632-3