Abstract
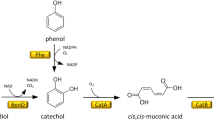
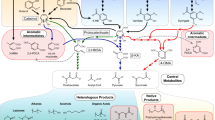
Microbial production of cis,cis-muconate (ccMA) from phenolic compounds obtained by chemical depolymerization of lignin is a promising approach to valorize lignin. Because microbial production requires a large amount of carbon and energy source, it is desirable to establish a ccMA-producing strain that utilizes lignin-derived phenols instead of general sources like glucose. We isolated Pseudomonas sp. strain NGC7 that grows well on various phenolic compounds derived from p-hydroxyphenyl, guaiacyl, and syringyl units of lignin. An NGC7 mutant of protocatechuate (PCA) 3,4-dioxygenase and ccMA cycloisomerase genes (NGC703) lost the ability to grow on vanillate and p-hydroxybenzoate but grew normally on syringate. Introduction of a plasmid carrying genes encoding PCA decarboxylase, flavin prenyltransferase, vanillate O-demethylase, and catechol 1,2-dioxygenase into NGC703 enabled production of 3.2 g/L ccMA from vanillate with a yield of 75% while growing on syringate. This strain also produced ccMA from birch lignin-derived phenols. All these results indicate the utility of NGC7 in glucose-free ccMA production.





Similar content being viewed by others
References
Abe T, Masai E, Miyauchi K, Katayama Y, Fukuda M (2005) A tetrahydrofolate-dependent O-demethylase, LigM, is crucial for catabolism of vanillate and syringate in Sphingomonas paucimobilis SYK-6. J Bacteriol 187:2030–2037. https://doi.org/10.1128/JB.187.6.2030-2037.2005
Barton N, Horbal L, Starck S, Kohlstedt M, Luzhetskyy A, Wittmann C (2018) Enabling the valorization of guaiacol-based lignin: integrated chemical and biochemical production of cis,cis-muconic acid using metabolically engineered Amycolatopsis sp. ATCC 39116. Metab Eng 45:200–210. https://doi.org/10.1016/j.ymben.2017.12.001
Becker J, Kuhl M, Kohlstedt M, Starck S, Wittmann C (2018) Metabolic engineering of Corynebacterium glutamicum for the production of cis,cis-muconic acid from lignin. Microb Cell Fact 17:115. https://doi.org/10.1186/s12934-018-0963-2
Beckham GT, Johnson CW, Karp EM, Salvachua D, Vardon DR (2016) Opportunities and challenges in biological lignin valorization. Curr Opin Biotechnol 42:40–53. https://doi.org/10.1016/j.copbio.2016.02.030
Blatny JM, Brautaset T, Winther-Larsen HC, Karunakaran P, Valla S (1997) Improved broad-host-range RK2 vectors useful for high and low regulated gene expression levels in gram-negative bacteria. Plasmid 38:35–51. https://doi.org/10.1006/plas.1997.1294
Boerjan W, Ralph J, Baucher M (2003) Lignin biosynthesis. Annu Rev Plant Biol 54:519–546. https://doi.org/10.1146/annurev.arplant.54.031902.134938
Burlage RS, Hooper SW, Sayler GS (1989) The TOL (pWW0) catabolic plasmid. Appl Environ Microbiol 55:1323
Cecil JH, Garcia DC, Giannone RJ, Michener JK (2018) Rapid, parallel identification of catabolism pathways of lignin-derived aromatic compounds in Novosphingobium aromaticivorans. Appl Environ Microbiol 84:e01185-18. https://doi.org/10.1128/AEM.01185-18
Crawford RL (1975) Novel pathway for degradation of protocatechuic acid in Bacillus species. J Bacteriol 121:531–536
Das A, Rahimi A, Ulbrich A, Alherech M, Motagamwala AH, Bhalla A, da Costa Sousa L, Balan V, Dumesic JA, Hegg EL, Dale BE, Ralph J, Coon JJ, Stahl SS (2018) Lignin conversion to low-molecular-weight aromatics via an aerobic oxidation-hydrolysis sequence: comparison of different lignin sources. ACS Sustain Chem Eng 6:3367–3374. https://doi.org/10.1021/acssuschemeng.7b03541
Harwood CS, Parales RE (1996) The β-ketoadipate pathway and the biology of self-identity. Annu Rev Microbiol 50:553–590
Jiménez JI, Miñambres B, García JL, Díaz E (2002) Genomic analysis of the aromatic catabolic pathways from Pseudomonas putida KT2440. Environ Microbiol 4:824–841. https://doi.org/10.1046/j.1462-2920.2002.00370.x
Johnson CW, Abraham PE, Linger JG, Khanna P, Hettich RL, Beckham GT (2017) Eliminating a global regulator of carbon catabolite repression enhances the conversion of aromatic lignin monomers to muconate in Pseudomonas putida KT2440. Metab Eng Commun 5:19–25. https://doi.org/10.1016/j.meteno.2017.05.002
Johnson CW, Salvachua D, Khanna P, Smith H, Peterson DJ, Beckham GT (2016) Enhancing muconic acid production from glucose and lignin-derived aromatic compounds via increased protocatechuate decarboxylase activity. Metab Eng Commun 3:111–119. https://doi.org/10.1016/j.meteno.2016.04.002
Johnson M, Zaretskaya I, Raytselis Y, Merezhuk Y, McGinnis S, Madden TL (2008) NCBI BLAST: a better web interface. Nucleic Acids Res 36:W5–W9. https://doi.org/10.1093/nar/gkn201
Kamimura N, Masai E (2014) The protocatechuate 4,5-cleavage pathway: overview and new findings. In: Nojiri H, Tsuda M, Fukuda M, Kamagata Y (eds) Biodegradative bacteria: how bacteria degrade, survive, adapt, and evolve. Springer, Tokyo, pp 207–226. https://doi.org/10.1007/978-4-431-54520-0_10
Kasai D, Fujinami T, Abe T, Mase K, Katayama Y, Fukuda M, Masai E (2009) Uncovering the protocatechuate 2,3-cleavage pathway genes. J Bacteriol 191:6758–6768. https://doi.org/10.1128/JB.00840-09
Kasai D, Kamimura N, Tani K, Umeda S, Abe T, Fukuda M, Masai E (2012) Characterization of FerC, a MarR-type transcriptional regulator, involved in transcriptional regulation of the ferulate catabolic operon in Sphingobium sp. strain SYK-6. FEMS Microbiol Lett 332:68–75. https://doi.org/10.1111/j.1574-6968.2012.02576.x
Kasai D, Masai E, Miyauchi K, Katayama Y, Fukuda M (2005) Characterization of the gallate dioxygenase gene: three distinct ring cleavage dioxygenases are involved in syringate degradation by Sphingomonas paucimobilis SYK-6. J Bacteriol 187:5067–5074. https://doi.org/10.1128/JB.187.15.5067-5074.2005
Maruyama K, Ariga N, Tsuda M, Deguchi K (1978) Purification and properties of α-hydroxy-γ-carboxymuconic ε-semialdehyde dehydrogenase. J Biochem 83:1125–1134
Masai E, Katayama Y, Fukuda M (2007) Genetic and biochemical investigations on bacterial catabolic pathways for lignin-derived aromatic compounds. Biosci Biotechnol Biochem 71:1–15
Masai E, Sasaki M, Minakawa Y, Abe T, Sonoki T, Miyauchi K, Katayama Y, Fukuda M (2004) A novel tetrahydrofolate-dependent O-demethylase gene is essential for growth of Sphingomonas paucimobilis SYK-6 with syringate. J Bacteriol 186:2757–2765
Masai E, Shinohara S, Hara H, Nishikawa S, Katayama Y, Fukuda M (1999) Genetic and biochemical characterization of a 2-pyrone-4,6-dicarboxylic acid hydrolase involved in the protocatechuate 4,5-cleavage pathway of Sphingomonas paucimobilis SYK-6. J Bacteriol 181:55–62
Nogales J, Canales A, Jimenez-Barbero J, Garcia JL, Diaz E (2005) Molecular characterization of the gallate dioxygenase from Pseudomonas putida KT2440. The prototype of a new subgroup of extradiol dioxygenases. J Biol Chem 280:35382–35390
Nozaki M, Kagamiyama H, Hayaishi O (1963) Metapyrocatechase I. Purification, crystallization and some properties. Biochemische Zeitschrift 338:582–590
Payer SE, Marshall SA, Barland N, Sheng X, Reiter T, Dordic A, Steinkellner G, Wuensch C, Kaltwasser S, Fisher K, Rigby SEJ, Macheroux P, Vonck J, Gruber K, Faber K, Himo F, Leys D, Pavkov-Keller T, Glueck SM (2017) Regioselective para-carboxylation of catechols with a prenylated flavin dependent decarboxylase. Angew Chem Int Ed Engl 56:13893–13897. https://doi.org/10.1002/anie.201708091
Ragauskas AJ, Beckham GT, Biddy MJ, Chandra R, Chen F, Davis MF, Davison BH, Dixon RA, Gilna P, Keller M, Langan P, Naskar AK, Saddler JN, Tschaplinski TJ, Tuskan GA, Wyman CE (2014) Lignin valorization: improving lignin processing in the biorefinery. Science 344:1246843. https://doi.org/10.1126/science.1246843
Ralph J, Lundquist K, Brunow G, Lu F, Kim H, Schatz PF, Marita JM, Hatfield RD, Ralph SA, Christensen JH, Boerjan W (2004) Lignins: natural polymers from oxidative coupling of 4-hydroxyphenyl-propanoids. Phytochem Rev 3:29–60. https://doi.org/10.1023/B:PHYT.0000047809.65444.a4
Ravi K, Garcia-Hidalgo J, Gorwa-Grauslund MF, Liden G (2017) Conversion of lignin model compounds by Pseudomonas putida KT2440 and isolates from compost. Appl Microbiol Biotechnol 101:5059–5070. https://doi.org/10.1007/s00253-017-8211-y
Rice P, Longden I, Bleasby A (2000) EMBOSS: the european molecular biology open software suite. Trends Genet 16:276–277
Rodriguez A, Salvachúa D, Katahira R, Black BA, Cleveland NS, Reed M, Smith H, Baidoo EEK, Keasling JD, Simmons BA, Beckham GT, Gladden JM (2017) Base-catalyzed depolymerization of solid lignin-rich streams enables microbial conversion. ACS Sustain Chem Eng 5:8171–8180. https://doi.org/10.1021/acssuschemeng.7b01818
Salvachúa D, Johnson CW, Singer CA, Rohrer H, Peterson DJ, Black BA, Knapp A, Beckham GT (2018) Bioprocess development for muconic acid production from aromatic compounds and lignin. Green Chem 20:5007–5019. https://doi.org/10.1039/c8gc02519c
Sarkanen KV, Ludwig CH (1971) Lignins: occurrence, formation, structure and reactions. Wiley, New York
Schäfer A, Tauch A, Jäger W, Kalinowski J, Thierbach G, Pühler A (1994) Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene 145:69–73
Shikinaka K, Otsuka Y, Nakamura M, Masai E, Katayama Y (2018) Utilization of lignocellulosic biomass via novel sustainable process. J Oleo Sci 67:1059–1070. https://doi.org/10.5650/jos.ess18075
Sonoki T, Morooka M, Sakamoto K, Otsuka Y, Nakamura M, Jellison J, Goodell B (2014) Enhancement of protocatechuate decarboxylase activity for the effective production of muconate from lignin-related aromatic compounds. J Biotechnol 192 Pt A:71–77. https://doi.org/10.1016/j.jbiotec.2014.10.027
Sonoki T, Takahashi K, Sugita H, Hatamura M, Azuma Y, Sato T, Suzuki S, Kamimura N, Masai E (2017) Glucose-free cis, cis-muconic acid production via new metabolic designs corresponding to the heterogeneity of lignin. ACS Sustain Chem Eng 6:1256–1264. https://doi.org/10.1021/acssuschemeng.7b03597
Sugimoto K, Senda M, Kasai D, Fukuda M, Masai E, Senda T (2014) Molecular mechanism of strict substrate specificity of an extradiol dioxygenase, DesB, derived from Sphingobium sp. SYK-6. PLoS One 9:e92249. https://doi.org/10.1371/journal.pone.0092249
Vanholme R, Demedts B, Morreel K, Ralph J, Boerjan W (2010) Lignin biosynthesis and structure. Plant Physiol 153:895–905. https://doi.org/10.1104/pp.110.155119
Vardon DR, Franden MA, Johnson CW, Karp EM, Guarnieri MT, Linger JG, Salm MJ, Strathmann TJ, Beckham GT (2015) Adipic acid production from lignin. Energy Environ Sci 8:617–628. https://doi.org/10.1039/c4ee03230f
White MD, Payne KA, Fisher K, Marshall SA, Parker D, Rattray NJ, Trivedi DK, Goodacre R, Rigby SE, Scrutton NS, Hay S, Leys D (2015) UbiX is a flavin prenyltransferase required for bacterial ubiquinone biosynthesis. Nature 522:502–506. https://doi.org/10.1038/nature14559
Williams PA, Sayers JR (1994) The evolution of pathways for aromatic hydrocarbon oxidation in Pseudomonas. Biodegradation 5:195–217. https://doi.org/10.1007/BF00696460
Wu W, Dutta T, Varman AM, Eudes A, Manalansan B, Loque D, Singh S (2017) Lignin valorization: two hybrid biochemical routes for the conversion of polymeric lignin into value-added chemicals. Sci Rep 7:8420. https://doi.org/10.1038/s41598-017-07895-1
Xu Z, Lei P, Zhai R, Wen Z, Jin M (2019) Recent advances in lignin valorization with bacterial cultures: microorganisms, metabolic pathways, and bio-products. Biotechnol Biofuels. https://doi.org/10.1186/s13068-019-1376-0
Acknowledgements
We thank the Advanced Low Carbon Technology Development (ALCA) program, Japan Science and Technology Agency (JPMJAL1506) for funding.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
N.K, T.S, and E.M. are inventors on a patent related to this work. The authors declare that they have no other conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Shinoda, E., Takahashi, K., Abe, N. et al. Isolation of a novel platform bacterium for lignin valorization and its application in glucose-free cis,cis-muconate production. J Ind Microbiol Biotechnol 46, 1071–1080 (2019). https://doi.org/10.1007/s10295-019-02190-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10295-019-02190-6