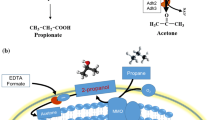
Abstract
Propane is the major component of liquefied petroleum gas (LPG). Nowadays, the use of LPG is decreasing, and thus utilization of propane as a chemical feedstock is in need of development. An efficient biological conversion of propane to acetone using a methanotrophic whole cell as the biocatalyst was proposed and investigated. A bio-oxidation pathway of propane to acetone in Methylomonas sp. DH-1 was analyzed by gene expression profiling via RNA sequencing. Propane was oxidized to 2-propanol by particulate methane monooxygenase and subsequently to acetone by methanol dehydrogenases. Methylomonas sp. DH-1 was deficient in acetone-converting enzymes and thus accumulated acetone in the absence of any enzyme inhibition. The maximum accumulation, average productivity and specific productivity of acetone were 16.62 mM, 0.678 mM/h and 0.141 mmol/g cell/h, respectively, under the optimized conditions. Our study demonstrates a novel method for the bioconversion of propane to acetone using methanotrophs under mild reaction condition.





Similar content being viewed by others
References
Althuluth M, Mota-Martinez MT, Berrouk A, Kroon MC, Peters CJ (2014) Removal of small hydrocarbons (ethane, propane, butane) from natural gas streams using the ionic liquid 1-ethyl-3-methylimidazolium tris (pentafluoroethyl) trifluorophosphate. J Supercrit Fluids 90:65–72
Baker RW, Lokhandwala K (2008) Natural gas processing with membranes: an overview. Ind Eng Chem Res 47:2109–2121
Burrows KJ, Cornish A, Scott D, Higgins IJ (1984) Substrate specificities of the soluble and particulate methane mono-oxygenases of Methylosinus trichosporium OB3b. Microbiol 130:3327–3333
Crombie AT, Murrell JC (2014) Trace-gas metabolic versatility of the facultative methanotroph Methylocella silvestris. Nat 510:148–151
Elliott SJ, Zhu M, Tso L, Nguyen HHT, Yip JHK, Chan SI (1997) Regio-and stereoselectivity of particulate methane monooxygenase from Methylococcus capsulatus (Bath). J Am Chem Soc 119:9949–9955
Ezeji TC, Qureshi N, Blaschek HP (2003) Production of acetone, butanol and ethanol by Clostridium beijerinckii BA101 and in situ recovery by gas stripping. World J Microbiol Biotechnol 19:595–603
Hanson RS, Hanson TE (1996) Methanotrophic bacteria. Microbiol Rev 60:439–471
Hou CT, Patel RN, Laskin AI, Barnabe N, Marczak I (1979) Identification and purification of a nicotinamide adenine dinucleotide-dependent secondary alcohol dehydrogenase from C1-utilizing microbes. FEBS Lett 101:179–183
Hur DH, Na JG, Lee EY (2016) Highly efficient bioconversion of methane to methanol using a novel type I Methylomonas sp. DH‐1 newly isolated from brewery waste sludge. J Chem Technol Biotechnol. doi:10.1002/jctb.5007
Hwang IY, Hur DH, Lee JH, Park CH, Chang IS, Lee JW, Lee EY (2015) Batch conversion of methane to methanol using Methylosinus trichosporium OB3b as biocatalyst. J Microbiol Biotechnol 25:375–380
Itoh S, Kawakami H, Fukuzumi S (1998) Model studies on calcium-containing quinoprotein alcohol dehydrogenases. catalytic role of Ca2+ for the oxidation of alcohols by coenzyme PQQ (4,5-Dihydro-4,5-dioxo-1H-pyrrolo[2,3-f]quinoline-2,7,9-tricarboxylic Acid). Biochemistry 37:6562–6571
Itoh S, Kawakami H, Fukuzumi S (1997) Modeling of the chemistry of quinoprotein methanol dehydrogenase. oxidation of methanol by calcium complex of coenzyme PQQ via addition–elimination mechanism. J Am Chem Soc 119:439–440
Kang TJ, Lee EY (2016) Metabolic versatility of microbial methane oxidation for biocatalytic methane conversion. J Ind Eng Chem 35:8–13
Koop DR (1992) Oxidative and reductive metabolism by cytochrome P450 2E1. Faseb J 6:724–730
Kotani T, Kawashima Y, Yurimoto H, Kato N, Sakai Y (2006) Gene structure and regulation of alkane monooxygenases in propane-utilizing Mycobacterium sp. TY-6 and Pseudonocardia sp. TY-7. J Biosci Bioeng 102:184–192
Kotani T, Yurimoto H, Kato N, Sakai Y (2007) Novel acetone metabolism in a propane-utilizing bacterium, Gordonia sp. strain TY-5. J Bacteriol 189:886–893
Kumar S, Kwon HT, Choi KH, Lim W, Cho JH, Tak K, Moon I (2011) LNG: an eco-friendly cryogenic fuel for sustainable development. Appl Energy 88:4264–4273
Miyazaki SS, Izumi Y, Yamada H (1987) Purification and characterization of methanol dehydrogenase of a serine-producing methylotroph, Hyphomicrobium methylovorum. J Ferment Bioeng 65:371–377
NGVAMERICA (2014) A comparison of compressed natural gas and propane. http://www.ngvamerica.org/pdfs/CNG_LPG_4.23.14.pdf. Accessed 26 Dec 2016
Patel RN, Hou CT, Laskin AI, Felix A (1982) Microbial oxidation of hydrocarbons: properties of a soluble methane monooxygenase from a facultative methane-utilizing organism, Methylobacterium sp. strain CRL-26. Appl Environ Microbiol 44:1130–1137
Patel SK, Mardina P, Kim SY, Lee JK, Kim IW (2016) Biological methanol production by a type ii methanotroph Methylocystis bryophila. J Microbiol Biotechnol. doi:10.4014/jmb.1601.01013
Patel SK, Selvaraj C, Mardina P, Jeong JH, Kalia VC, Kang YC, Lee JK (2016) Enhancement of methanol production from synthetic gas mixture by Methylosinus sporium through covalent immobilization. Appl Energy 171:383–391
Sifniades S, Levy AB, Bahl H (2010) Acetone. Ullmann’s encyclopedia of industrial chemistry. doi:10.1002/14356007.a01_079.pub2
Stephens GM, Dalton H (1986) The role of the terminal and subterminal oxidation pathways in propane metabolism by bacteria Microbiol 132:2453–2462
US Department of Energy (2015) CLEAN CITIES alternative fuel price report. http://www.afdc.energy.gov/uploads/publication/alternative_fuel_price_report_october_2015.pdf. Accessed 26 Dec 2016
Van Beilen JB, Funhoff EG (2007) Alkane hydroxylases involved in microbial alkane degradation. Appl Microbiol Biotechnol 74:13–21
Vanderberg LA, Perry JJ (1994) Dehalogenation by Mycobacterium vaccae JOB-5: role of the propane monooxygenase. Can J Microbiol 40:169–172
Weiner ML, Kotkoskie LA (1999) Excipient toxicity and safety. Marcel Dekker, New York
Acknowledgements
This research was supported by the C1 Gas Refinery Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Science, ICT and Future Planning (2015M3D3A1A01064882).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Hur, D.H., Nguyen, T.T., Kim, D. et al. Selective bio-oxidation of propane to acetone using methane-oxidizing Methylomonas sp. DH-1. J Ind Microbiol Biotechnol 44, 1097–1105 (2017). https://doi.org/10.1007/s10295-017-1936-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10295-017-1936-x