Abstract
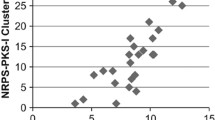
Actinomycetes are historically important sources for secondary metabolites (SMs) with applications in human medicine, animal health, and plant crop protection. It is now clear that actinomycetes and other microorganisms with large genomes have the capacity to produce many more SMs than was anticipated from standard fermentation studies. Indeed ~90 % of SM gene clusters (SMGCs) predicted from genome sequencing are cryptic under conventional fermentation and analytical analyses. Previous studies have suggested that among the actinomycetes with large genomes, some have the coding capacity to produce many more SMs than others, and that strains with the largest genomes tend to be the most gifted. These contentions have been evaluated more quantitatively by antiSMASH 3.0 analyses of microbial genomes, and the results indicate that many actinomycetes with large genomes are gifted for SM production, encoding 20–50 SMGCs, and devoting 0.8–3.0 Mb of coding capacity to SM production. Several Proteobacteria and Firmacutes with large genomes encode 20–30 SMGCs and devote 0.8–1.3 Mb of DNA to SM production, whereas cultured bacteria and archaea with small genomes devote insignificant coding capacity to SM production. Fully sequenced genomes of uncultured bacteria and archaea have small genomes nearly devoid of SMGCs.

Similar content being viewed by others
References
Aigle B, Lautru S, Spiteller D, Dickschat JS, Challis GL, Leblond P, Pernodet JL (2014) Genome mining of Streptomyces ambofaciens. J Ind Microbiol Biotechnol 41:251–264
Albertsen M, Hugenholtz P, Skarsheweski A, Nielsen KL, Tyson GW, Nielsen PH (2013) Genome sequences of rare, uncultured bacteria obtained by differential coverage binning of multiple metagenomes. Nat Biotechnol 31:533–538
Amann RI, Ludwig W, Schleifer KH (1995) Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev 59:143–169
Anantharaman K, Brown CT, Burstein D, Castelle CJ, Probst AJ, Thomas BC, Williams KH, Banfield JF (2016) Analysis of five complete genome sequences for members of the class Perigrinibacteria bacterial phylum. Peer J 4:e1607
Bachmann BO, Ravel J (2009) Chapter 8. Methods for in silico prediction of microbial polyketide and nonribosomal peptide biosynthetic pathways from DNA sequence data. Methods Enzymol 458:181–217
Bachmann BO, Van Lanen SG, Baltz RH (2014) Microbial genome mining for accelerated natural products discovery: is a renaissance in the making? J Ind Microbiol Biotechnol 41:175–184
Baltz RH (2005) Antibiotic discovery from actinomycetes: will a renaissance follow the decline and fall? SIM News 55:186–196
Baltz RH (2007) Antimicrobials from actinomycetes; back to the future. Microbe 2:125–131
Baltz RH (2008) Renaissance in antibacterial discovery from actinomycetes. Curr Opin Pharmacol 8:557–563
Baltz RH (2011) Strain improvement in actinomycetes in the postgenomic era. J Ind Microbiol Biotechnol 38:657–666
Baltz RH (2011) Function of MbtH homologs in nonribosomal peptide biosynthesis and applications in secondary metabolite discovery. J Ind Microbiol Biotechnol 38:1747–1760
Baltz RH (2014) MbtH homology codes to identify gifted microbes for genome mining. J Ind Microbiol Biotechnol 41:357–369
Baltz RH (2016) Genetic manipulation of secondary metabolite biosynthesis for improved production in Streptomyces and other actinomycetes. J Ind Microbiol Biotechnol 43:343–370
Banik JJ, Brady SF (2008) Cloning and characterization of new glycopeptide gene clusters found in an environmental DNA megalibrary. Proc Nat Acad Sci USA 105:17273–17277
Banik JJ, Craig JW, Calle PY, Brady SF (2010) Tailoring enzyme-rich environmental DNA clones: a source of enzymes for generating libraries of unnatural natural products. J Am Chem Soc 132:15661–15670
Baranasic D, Gacesa R, Starcevic A et al (2013) Draft genome sequence of Streptomyces rapamycinicus strain NRRL 5491, the producer of the immunosuppressant rapamycin. Genome Announc 1:e00581-13
Barka EA, Vatsa P, Sanchez L et al (2016) Taxonomy, physiology, and natural products of Actinobacteria. Microbiol Mol Biol Rev 80:1–43
Bentley SD, Chater KF, Cerdeno-Tarraga AM et al (2002) Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature 417:141–147
Berdy J (2005) Bioactive microbial metabolites. J Antibiot 58:1–26
Blin K, Medema MH, Kazempour D, Fischbach MA, Breitling R, Takano E, Weber T (2013) antiSMASH 2.0—a versatile platform for genome mining of secondary metabolite producers. Nucleic Acids Res 41:W204–W212
Boddy CN (2014) Bioinformatics tools for genome mining of polyketide and non-ribosomal peptides. J Ind Microbiol Biotechnol 41:443–450
Brown CT, Hug LA, Thomas BC et al (2015) Unusual biology across a group comprising more than 15% of domain Bacteria. Nature 523:208–211
Butler MS, Robertson AA, Cooper MA (2014) Natural product and natural product derived drugs in clinical trials. Nat Prod Rep 31:1612–1661
Calteau A, Fewer DP, Latifi A et al (2014) Phylum-wide comparative genomics unravel the diversity of secondary metabolism in Cyanobacteria. BMC Genom 15:997
Challis GL (2014) Exploitation of the Streptomyces coelicolor A3(2) genome sequence for discovery of new natural products and biosynthetic pathways. J Ind Microbiol Biotechnol 41:219–232
Charlop-Powers Z, Owen JG, Reddy BVB, Ternei MA, Guimaraes DO, de Frias UA, Pupo MT, Seepe P, Feng Z, Brady SF (2015) Global biogeographic sampling of bacterial secondary metabolism. Elife 4:e05048
da Silva RR, Dorrestein PC, Quinn RA (2015) Illuminating the dark matter in metabolomics. Proc Nat Acad Sci USA 112:12549–12550
Demain AL (2014) Importance of microbial natural products and the need to revitalize their discovery. J Ind Microbiol Biotechnol 41:185–201
Donadio S, Monciardini P, Socio M (2007) Polyketide synthases and nonribosomal peptide synthases: the emerging view from bacterial genomics. Nat Prod Rep 24:1073–1109
Doroghazi JR, Metcalf WW (2013) Comparative genomics of actinomycetes with a focus on natural product biosynthetic genes. BMC Genom 14:611
Doroghazi JR, Albright JC, Goering AW et al (2014) A roadmap for natural product discovery based on large-scale genomics and metabolomics. Nat Chem Biol 10:963–968
Duchaud E, Rusnioc C, Franguel L et al (2003) The genome sequence of the entomopathogenic bacterium Photorhabdus luminescens. Nat Biotechnol 21:1307–1313
Dupont CL, Rusch DB, Yooseph S et al (2012) Genomic insight into SAR86, an abundant and uncultivated marine bacterial lineage. ISME 6:1186–1199
Fernandez-Martinez L, Borsetto C, Gomez-Escribano JP et al (2014) New insights into chloramphenicol biosynthesis in Streptomyces venezuelae ATCC 10712. Antimicrob Agents Chemother 58:7441–7450
Galm U, Sparks TC (2016) Natural product derived insecticides: discovery and development of spinetoram. J Ind Microbiol Biotechnol 43:185–193
Giddings LA, Newman DJ (2013) Microbial natural products: molecular blueprints for antitumor agents. J Ind Microbiol Biotechnol 40:1181–1210
Goering AW, McClure RA, Doroghazi JR et al (2016) Metabolgenomics: correlation of microbial gene clusters with metabolites drives discovery of a nonribosomal peptide with an unusual amino acid monomer. ACS Cent Sci 2:99–108
Goldman BS, Nierman WC, Kaiser D et al (2006) Evolution of sensory complexity recorded in a myxobacterial genome. Proc Nat Adad Sci USA 103:15200–15205
Hadjithomas M, Chen IM, Chu K et al (2015) IMG-ABC: a knowledge base to fuel discovery of biosynthetic gene clusters and novel secondary metabolites. MBio 6:e00932-15
Haroon MF, Thompson LR, Stingl U (2016) Draft genome of uncultured SAR324 bacterium lautmerah10, binned from Red Sea metagenome. Genome Announc 4:e01711–15
Hess M, Scayrba A, Egan R et al (2011) Metagenomic discovery of biomass-degrading genes and genomes from cow rumen. Science 331:463–467
Iftime D, Kulik A, Härtner T et al (2016) Identification and activation of novel biosynthetic gene clusters by genome mining in the kirromycin producer Tü 365. J Ind Microbiol Biotechnol 43:277–291
Ikeda H, Ishikawa J, Hanamoto A, Shinose M, Kikuchi H, Shiba T, Sakaki Y, Hattori M, Ōmura S (2003) Complete genome sequence and comparative analysis of the industrial microorganism Streptomyces avermitilis. Nat Biotechnol 21:526–531
Ikeda H, Shin-ya K, Ōmura S (2014) Genome mining of the Streptomyces avermitilis genome and development of genome-minimized hosts for heterologous expression of biosynthetic gene clusters. J Ind Microbiol Biotechnol 41:233–250
Isikawa J, Yamashita A, Mikami Y et al (2004) The complete genomic sequence of Nocardia farcinica IFM 10152. Proc Nat Acad Sci USA 101:14925–14930
Iversen V, Morris RM, Frazar CD, Berthiaume CT, Morales RL, Armbrust EV (2012) Untangling genomes from metagenomes: revealing an uncultured class of marine Eurarchaeota. Science 335:587–590
Johnston CW, Connaty AD, Skinnider MA, Li Y, Grunwald A, Wyatt MA, Kerr RG, Magarvey NA (2016) Informatic search strategies to discover analogues and variants of natural product archtypes. J Ind Microbiol Biotechnol 43:293–298
Kantor RS, Wrighton KC, Handley KM, Sharon I, Hug LA, Castelle CJ, Thomas BC, Banfield JF (2013) Small genomes and sparse metabolisms of sediment-associated bacteria from candidate phyla. MBio 4:e00708-13
Katz L, Baltz RH (2016) Natural product discovery: past, present, and future. J Ind Microbiolol Biotechnol 43:155–176
Katz M, Hover BM, Brady SF (2016) Culture-independent discovery of natural products from soil metagenomes. J Ind Microbiol Biotechnol 43:129–141
Knight-Connoni V, Mascio C, Chesnel L, Silverman J (2016) Discovery and development of surotomycin for treatment of Clostridium dificile. J Ind Microbiol Biotechnol 43:195–204
Kwun MJ, Hong HJ (2014) Genome sequence of Streptomyces toyocaensis NRRL 15009, producer of the glycopeptide antibiotic A47934. Genome Announc 2:e00749-14
Land M, Lapidus A, Mayilraj S et al (2009) Complete genome sequence of Actinosynnema mirum type strain (101). Stand Genomic Sci 1:46–53
Li YF, Tsai KJ, Harvey CJ et al (2016) Comprehensive curation and analysis of fungal biosynthetic gene clusters of published natural products. Fungal Genet Biol 89:18–28
Li ZF, Li X, Liu H et al (2011) Genome sequence of the halotolerant marine bacterium Myxococcus fulvus HW-1. J Bacteriol 193:5015–5016
Liolios K, Sikorski J, Jando M et al (2010) Complete genome sequence of Thermobispora bispora type strain (R51T). Stand Genomic Sci 2:318–326
Liu X, Cheng YQ (2014) Genome-guided discovery of diverse natural products from Burkholderia sp. J Ind Microbiol Biotechnol 41:275–284
Lykidis A, Mavromatis K, Ivanova N et al (2007) Genome sequence of the soil cellulolytic actinomycete Thermobifida fusca YX. J Bacteriol 189:2477–2486
Ma M, Wang Z, Li L et al (2012) Complete genome sequence of Paenibacillus mucilaginosus 3016, a bacterium functional as microbial fertilizer. J Bacteriol 194:2777–2778
Maffioli SI, Cruz JC, Monciardini P, Sosio M, Donadio S (2016) Advancing cell wall inhibitors towards clinical applications. J Ind Microbiol Biotechnol 43:177–187
Miao V, Coeffet-LeGal MF, Brian P et al (2005) Daptomycin biosynthesis in Streptomyces roseosporus: cloning and analysis of the gene cluster and revision of peptide stereochemistry. Microbiology 151:1507–1523
Moss NA, Bertin MJ, Kleigrewe K, Leao TF, Gerwick L, Gerwick WH (2016) Integrating mass spectrometry and genomics for cyanobacterial metabolite discovery. J Ind Microbiol Biotechnol 43:313–324
Narasingarao P, Podell S, Ugalde JA, Brochier C, Emerson JB, Brocks JJ, Heidelberg KB, Banfield JF, Allen EE (2012) De novo metagenomic assembly reveals abundant novel major lineage of Archaea in hypersaline microbial communities. ISME J 6:81–93
Newman DJ, Cragg GM (2016) Natural products as sources of new drugs from 1981 to 2014. J Nat Prod 79:629–661
Nolan M, Sikorski J, Jando M (2010) Complete genome sequence of Streptosporangium roseum type strain (NI 9100). Stand Genomic Sci 2:29–37
Ochi K, Hosaka T (2013) New strategies for drug discovery: activation of silent or weakly expressed microbial gene clusters. Appl Microbiol Biotechnol 97:87–98
Ochi K, Tanaka Y, Tojo S (2014) Activating the expression of bacterial cryptic genes by rpoB mutations in RNA polymerase or by rare earth elements. J Ind Microbiol Biotechnol 41:403–414
Ohnishi Y, Ishikawa J, Hara H et al (2008) Genome sequence of the streptomycin-producing microorganism Streptomyces griseus IFO 13350. J Bacteriol 190:4050–4060
Oliynyk M, Samborskyy M, Lester JB et al (2007) Complete genome sequence of the erythromycin-producing bacterium Saccharopolyspora erythraea NRRL2338. Nat Biotechnol 25:447–453
Pace NR (2009) Mapping the tree of life: progress and prospects. Microbiol Mol Biol Rev 73:565–576
Pati A, Sikorski J, Nolan M et al (2009) Complete genome sequence of Saccharomonospora viridis type strain (P101T). Stand Genomic Sci 1:141–149
Quadri LE, Weinreb PH, Lei M, Nakano MM, Zuber P, Walsh CT (1998) Characterization of Sfp, a Bacillus subtilis phosphopantetheinyl transferase for peptidyl carrier protein domains in peptide synthetases. Biochemistry 37:1585–1595
Rebets Y, Tokovenko B, Luzhetskyy A et al (2014) Complete genome sequence of producer of the glycopeptide antibiotic aculeximycin Kutzneria albida DSM 43870, a representative of minor genus of Pseudonocardiaceae. BMC Genom 15:885
Rinke C, Schwientek P, Sczyrba A et al (2013) Insights into the phylogeny and coding potential of microbial dark matter. Nature 499:431–437
Rückert C, Szczepanowski R, Albertsmeier A et al (2013) Complete genome sequence of the kirromycin producer Streptomyces collinus Tu 365 consisting of a linear chromosome and two linear plasmids. J Biotechnol 168:739–740
Rutledge PJ, Challis GL (2015) Discovery of microbial natural products by activation of silent biosynthetic gene clusters. Nat Rev Microbiol 13:509–523
Sales CM, Mahendra S, Grostern A et al (2011) Genome sequence of the 1,4-dioxane-degrading Pseudonocardia dioxanivorans strain CB1190. J Bacteriol 193:4549–4550
Schneiker S, Perlova O, Kaiser O et al (2007) Complete genome sequence of the myxobacterium Sorangium cellulosum. Nat Biotechnol 25:1281–1289
Shih PM, Wu D, Latifi A et al (2013) Improving the coverage of the cyanobacterial phylum using diversity-driven genome sequencing. Proc Nat Acad Sci USA 110:1053–1058
Skinnider MA, Dejong CA, Rees PN, Johnston CW, Li H, Webster AL, Wyatt MA, Magarvey NA (2015) Genomes to natural products PRediction Informatics for Secondary Metabolomes (PRISM). Nucleic Acids Res 43:9645–9662
Smanski MJ, Schlatter DC, Kinkel LL (2016) Leveraging ecological theory to guide natural product discovery. J Ind Microbiol Biotechnol 43:115–128
Strobel T, Al-Dilaim A, Blom J et al (2012) Complete genome sequence of Saccharothrix espanaensis DSM 44229T and comparasin to the other completely sequenced Pseudonocardiaceae. BMC Genom 13:465
Tanaka Y, Kasahara K, Hirose Y, Murakami K, Kugimiya R, Ochi K (2013) Activation and products of the cryptic secondary metabolite biosynthetic gene clusters by rifampin resistance (rpoB) mutations in actinomycetes. J Bacteriol 195:2959–2970
Thaker MN, Wang W, Spanogionnopoulos P, Waglechner N, King AM, Medina R, Wright GD (2013) Identifying producers of antibacterial compounds by screening for antibiotic resistance. Nat Biotechnol 31:922–927
Thaker MN, Waglechner N, Wright GD (2014) Antibiotic resistance-mediated isolation of scaffold-specific natural product producers. Nat Protocol 9:1469–1479
Thibessard A, Haas D, Gerbaud C et al (2015) Complete genome sequence of Streptomyces ambofaciens ATCC 23877, the spiramycin producer. J Biotechnol 214:117–118
Tice H, Mayilraj S, Sims D et al (2010) Complete genome sequence of Nakamurella multipartita type strain (Y-104T). Stand Genomic Sci 2:168–175
Udwary DW, Zeigler L, Astolkar RN et al (2007) Genome sequencing reveals complex secondary metabolome in the marine actinomycete Salinispora tropica. Proc Nat Acad Sci USA 104:10376–10381
Van der Lee TA, Medema MH (2016) Computational strategies for genome-based natural product discovery and engineering in fungi. Fungal Genet Biol 89:29–36
Wang H, Fewer DP, Holm L, Rouhiainen L, Sivonen K (2014) Atlas of nonribosomal peptide and polyketide biosynthetic pathways reveals common occurrence of nonmodular enzymes. Proc Nat Acad Sci USA 111:9259–9264
Wang H, Sivonen K, Fewer DP (2015) Genomic insights into the distribution, genetic diversity and evolution of polyketide synthases and nonribosomal peptide synthetases. Curr Opin Genet Dev 35:79–85
Wang XJ, Yan YJ, Zhang B et al (2010) Genome sequence of the milbamycin-producing bacterium Streptomyces bingchenggensis. J Bacteriol 192:4526–4527
Weber T, Blin K, Duddela S et al (2015) antiSMASH 3.0 – a comprehensive resource for the genome mining of biosynthetic gene clusters. Nucleic Acids Res 43:W237–W243
Weber T, Kim HU (2016) The secondary metabolite bioinformatics portal: computational tools to facilitate synthetic biology of secondary metabolite production. Synth Syst Biotechnol 1:69–79
Wiemann P, Keller NP (2014) Strategies for mining fungal natural products. J Ind Microbiol Biotechnol 41:301–313
Wu H, Qu S, Lu C et al (2013) Genomic and transcriptomic insights into the thermo-regulated biosynthesis of validamycin in Streptomyces hygroscopicus 5008. BMC Genom 13:337
Xu L, Huang H, Wei W et al (2014) Complete genome sequence and comparative genomic analysis of the vancomycin-producing Amycolatopsis orientalis. BMC Genom 15:363
Yaegashi J, Oakley BR, Wang CC (2014) Recent advances in genome mining fungal natural products. J Ind Microbiol Biotechnol 41:433–442
Yamamura H, Ohnishi Y, Ishikawa J et al (2013) Complete genome sequence of the motile actinomyciete Actinoplanes missouriensis 431T (= NBRC 102363). Stand Genome Sci 7:294–303
Zaburannyi N, Rabyk M, Ostash B et al (2014) Insights into the naturally minimized Streptomyces albus J1074 gemone. BMC Genom 15:97
Zhao W, Zhong Y, Yuan H et al (2010) Complete genome sequence of the rifamycin SV-producing Amycolatopsis mediterranei U32 revealed its genetic characteristics in phylogeny and metabolism. Cell Res 20:1096–1108
Zhu F, Qin T, Tao L et al (2011) Clustered patterns of species origins of nature-derived drugs and clues for future bioprospecting. Proc Nat Acad Sci USA 31:12943–12948
Zhu H, Sandiford SK, van Wezel GP (2014) Triggers and cues that activate antibiotic production by actinomycetes. J Ind Microbiol Biotechnol 41:371–386
Acknowledgments
I would like to thank Arny Demain for his unfailing advocacy for natural products spanning many decades, and his many contributions to industrial biotechnology, starting with his seminal publications of cephalosporin C biosynthesis initiated during his tenure at Merck. It has been an honor to know Arny as a friend and mentor, and to contribute to this Special Issue of JIMB celebrating his 90th birthday! I would also like to acknowledge David Hopwood and Satoshi Ōmura and their colleagues for seminal observations on the cryptic secondary metabolite pathways encoded by S. coelicolor and S. avermitilis that have helped catalyze the resurgence of natural product research, particularly the genome mining approach discussed in this article.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Baltz, R.H. Gifted microbes for genome mining and natural product discovery. J Ind Microbiol Biotechnol 44, 573–588 (2017). https://doi.org/10.1007/s10295-016-1815-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10295-016-1815-x