Abstract
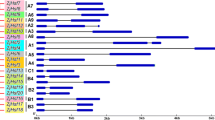
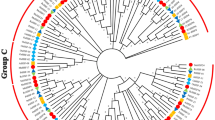
The heat stress transcription factors (Hsfs) play a prominent role in thermotolerance and eliciting the heat stress response in plants. Identification and expression analysis of Hsfs gene family members in chickpea would provide valuable information on heat stress responsive Hsfs. A genome-wide analysis of Hsfs gene family resulted in the identification of 22 Hsf genes in chickpea in both desi and kabuli genome. Phylogenetic analysis distinctly separated 12 A, 9 B, and 1 C class Hsfs, respectively. An analysis of cis-regulatory elements in the upstream region of the genes identified many stress responsive elements such as heat stress elements (HSE), abscisic acid responsive element (ABRE) etc. In silico expression analysis showed nine and three Hsfs were also expressed in drought and salinity stresses, respectively. Q-PCR expression analysis of Hsfs under heat stress at pod development and at 15 days old seedling stage showed that CarHsfA2, A6, and B2 were significantly upregulated in both the stages of crop growth and other four Hsfs (CarHsfA2, A6a, A6c, B2a) showed early transcriptional upregulation for heat stress at seedling stage of chickpea. These subclasses of Hsfs identified in this study can be further evaluated as candidate genes in the characterization of heat stress response in chickpea.









Similar content being viewed by others
References
Almoguera C, Rojas A, Díaz-Martín J, Prieto-Dapena P, Carranco R, Jordano J (2002) A seed-specific heat-shock transcription factor involved in developmental regulation during embryogenesis in sunflower. J Biol Chem 277:43866–43872
Baniwal SK, Bharti K, Chan KY, Fauth M, Ganguli A, Kotak S, Mishra SK, Nover L, Port M, Scharf KD, Tripp J, Weber C, Zielinski D, von Koskull-Döring P (2004) Heat stress response in plants: a complex game with chaperones and more than twenty heat stress transcription factors. J Biosci 29:471–487
Burge C, Karlin S (1997) Prediction of complete gene structures in human genomic DNA. J Mol Biol 268:78–94
Charng YY, Liu HC, Liu NY, Chi WT, Wang CN, Chang SH, Wang TT (2007) A heat-inducible transcription factor, HsfA2, is required for the extension of acquired thermotolerance in Arabidopsis. Plant Physiol 143:251–262
Chauhan H, Khurana N, Agarwal P, Khurana P (2011) Heat shock factors in rice (Oryza sativa L.): genome-wide expression analysis during reproductive development and abiotic stress. Mol Genet Genom 286:171–187
Chung E, Kim KM, Lee JH (2013) Genome-wide analysis and molecular characterization of heat shock transcription factor family in Glycine max. J Genet Genom 40:127–135
Cicero MP, Hubl ST, Harrison CJ, Littlefield O, Hardy JA, Nelson HCM (2001) The wing in yeast heat shock transcription factor (HSF) DNA-binding domain is required for full activity. Nucleic Acids Res 29:1715–1723
Dai C, Whitesell L, Rogers AB, Lindquist S (2007) Heat shock factor 1 is a powerful multifaceted modifier of carcinogenesis. Cell 130:1005–1018
Deokar A, Ramsay L, Sharpe AG, Diapari M, Sindhu A, Bett K, Warkentin TD, Tar’an B (2014) Genome-wide SNP identification in chickpea for use in the development of a high-density genetic map and improvement of the chickpea reference genome assembly. BMC Genom 15:708
Devasirvatham V, Gaur PM, Mallikarjuna N, Raju TN, Trethowan RM, Tan DKY (2013) Reproductive biology of chickpea response to heat stress in the field is associated with the performance in controlled environments. F Crop Res 142:9–19
Edgar RC (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797
Enoki Y, Sakurai H (2011) Diversity in DNA recognition by heat shock transcription factors (HSFs) from model organisms. FEBS Lett 585:1293–1298
Evrard A, Kumar M, Lecourieux D, Lucks J, von Koskull-Döring P, Hirt H (2013) Regulation of the heat stress response in Arabidopsis by MPK6-targeted phosphorylation of the heat stress factor HsfA2. PeerJ 1:e59
Finn RD, Bateman A, Clements J, Coggill P, Eberhardt RY, Eddy SR, Heger A, Hetherington K, Holm L, Mistry J, Sonnhammer ELL, Tate J, Punta M (2014) Pfam: the protein families database. Nucleic Acids Res 42:D222–D230
Fragkostefanakis S, Röth S, Schleiff E, Scharf KD (2015) Prospects of engineering thermotolerance in crops through modulation of heat stress transcription factor and heat shock protein networks. Plant Cell Environ 38:1881–1895
Garg R, Sahoo A, Tyagi AK, Jain M (2010) Validation of internal control genes for quantitative gene expression studies in chickpea (Cicer arietinum L.). Biochem Biophys Res Commun 396:283–288
Garg R, Patel RK, Jhanwar S, Priya P, Bhattacharjee A, Yadav G, Bhatia S, Chattopadhyay D, Tyagi AK, Jain M (2011) Gene discovery and tissue-specific transcriptome analysis in chickpea with massively parallel pyrosequencing and web resource development. Plant Physiol 156:1661–1678
Giorno F, Guerriero G, Baric S, Mariani C (2012) Heat shock transcriptional factors in Malus domestica: identification, classification and expression analysis. BMC Genom 13:639
Guo J, Wu J, Ji Q, Wang C, Luo L, Yuan Y, Wang Y, Wang J (2008) Genome-wide analysis of heat shock transcription factor families in rice and Arabidopsis. J Genet Genom 35:105–118
Hahn A, Bublak D, Schleiff E, Scharf KD (2011) Crosstalk between Hsp90 and Hsp70 chaperones and heat stress transcription factors in tomato. Plant Cell 23:741–755
Ikeda M, Mitsuda N, Ohme-Takagi M (2011) Arabidopsis HsfB1 and HsfB2b act as repressors of the expression of heat-inducible Hsfs but positively regulate the acquired thermotolerance. Plant Physiol 157:1243–1254
Jain M, Misra G, Patel RK, Priya P, Jhanwar S, Khan AW, Shah N, Singh VK, Garg R, Jeena G, Yadav M, Kant C, Sharma P, Yadav G, Bhatia S, Tyagi AK, Chattopadhyay D (2013) A draft genome sequence of the pulse crop chickpea (Cicer arietinum L.). Plant J 74:715–729
Jedlicka P, Mortin MA, Wu C (1997) Multiple functions of Drosophila heat shock transcription factor in vivo. EMBO J 16:2452–2462
Jin G-H, Gho H-J, Jung K-H (2013) A systematic view of rice heat shock transcription factor family using phylogenomic analysis. J Plant Physiol 170:321–329
Kotak S, Port M, Ganguli A, Bicker F, von Koskull-Döring P (2004) Characterization of C-terminal domains of Arabidopsis heat stress transcription factors (Hsfs) and identification of a new signature combination of plant class A Hsfs with AHA and NES motifs essential for activator function and intracellular localization. Plant J 39:98–112
Krishnamurthy L, Gaur PM, Basu PS, Chaturvedi SK, Tripathi S, Vadez V (2011) Large genetic variation for heat tolerance in the reference collection of chickpea (Cicer arietinum L.) germplasm. Plant Genet Res 9:1–26
Kumar M, Busch W, Birke H, Kemmerling B, Nürnberger T, Schöffl F (2009) Heat shock factors HsfB1 and HsfB2b are involved in the regulation of Pdf1.2 expression and pathogen resistance in Arabidopsis. Mol Plant 2:152–165.
Lescot M, Déhais P, Thijs G, Marchal K, Moreau Y, Van de Peer Y, Rombauts S (2002) PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res 30:325–327
Letunic I, Copley RR, Schmidt S, Ciccarelli FD, Doerks T, Schultz J, Ponting CP, Bork P (2004) SMART 4.0: towards genomic data integration. Nucleic Acids Res 32:D142–D144
Lin YX, Jiang HY, Chu ZX, Tang XL, Zhu SW, Cheng BJ (2011) Genome-wide identification, classification and analysis of heat shock transcription factor family in maize. BMC Genom 12:76
Lin Y, Cheng Y, Jin J, Jin X, Jiang H, Yan H, Cheng B (2014) Genome duplication and gene loss affect the evolution of heat shock transcription factor genes in legumes. PLoS One 9:e102825
Liu H, Charng Y (2013) Common and distinct functions of Arabidopsis class A1 and A2 heat shock factors in diverse abiotic stress responses and development. Plant Physiol 163:276–290
Lomsadze A, Ter-Hovhannisyan V, Chernoff Y, Borodovsky M (2005) Gene identification in novel eukaryotic genomes by self-training algorithm. Nucleic Acids Res 33:6494–6506
MapInspectsoftware (http://www.plantbreeding.wur.nl/UK/software_mapinspect.html) downloaded and analyzed on June, 2015
Maruyama KY, Todaka DA, Mizoi JU, Yoshida TA, Kidokoro SA, Matsukura SA, Takasaki HI, Sakurai TE, Yamamoto YOY, Yoshiwara KY (2012) Identification of Cis-acting promoter elements in cold- and dehydration-induced transcriptional pathways in Arabidopsis, rice, and soybean. DNA Res 19:37–49
Mishra SK, Tripp J, Winkelhaus S, Tschiersch B, Theres K, Nover L, Scharf K-D (2002) In the complex family of heat stress transcription factors, HsfA1 has a unique role as master regulator of thermotolerance in tomato. Genes Dev 16:1555–1567
Mistry J, Finn RD, Eddy SR, Bateman A, Punta M (2013) Challenges in homology search: HMMER3 and convergent evolution of coiled-coil regions. Nucleic Acids Res 41:e121–e121
Morley JF, Morimoto RI (2004) Regulation of longevity in Caenorhabditis elegans by heat shock factor and molecular chaperones. Mol Biol Cell 15:657–664
Mount DW (2009) Using hidden Markov models to align multiple sequences. Cold Spring Harb Protoc 2009:41
Nover L, Scharf KD, Gagliardi D, Vergne P, Czarnecka-Verner E, Gurley WB (1996) The Hsf world: classification and properties of plant heat stress transcription factors. Cell Stress Chaperones 1:215–238
Nover L, Bharti K, Döring P, Mishra SK, Ganguli A, Scharf K-D (2001) Arabidopsis and the heat stress transcription factor world: how many heat stress transcription factors do we need? Cell Stress Chaperones 6:177–189
Ogawa D, Yamaguchi K, Nishiuchi T (2007) High-level overexpression of the Arabidopsis HsfA2 gene confers not only increased thermotolerance but also salt/osmotic stress tolerance and enhanced callus growth. J Exp Bot 58:3373–3383
Parween S, Nawaz K, Roy R, Pole AK, Suresh BV, Misra G, Bhatia S (2015) An advanced draft genome assembly of a desi type chickpea (Cicer arietinum L.). Sci Rep 5:12806
Qin F, Kakimoto M, Sakuma Y, Maruyama K, Osakabe Y, Tran L-SP, Shinozaki K, Yamaguchi-Shinozaki K (2007) Regulation and functional analysis of ZmDREB2A in response to drought and heat stresses in Zea mays L. Plant J 50:54–69
Richards CL, Rosas U, Banta J, Bhambhra N, Purugganan MD (2012) Genome-wide patterns of Arabidopsis gene expression in nature. PLoS Genet 8:e1002662
Scharf KD, Berberich T, Ebersberger I, Nover L (2012) The plant heat stress transcription factor (Hsf) family: structure, function, and evolution. Biochim Biophys Acta 1819:104–119
Schultheiss J, Kunert O, Gase U, Scharf KD, Nover L, Rüterjans H (1996) Solution structure of the DNA-binding domain of the tomato heat stress transcription factor HSF24. Eur. J Biochem 236:911–921
Sharma R, Rawat V, Suresh CG (2014) Genome-Wide Identification and Tissue-Specific Expression Analysis of UDP-Glycosyltransferases Genes Confirm Their Abundance in Cicer arietinum (Chickpea) Genome. PLoS One 9:e109715
Soares-cavalcanti NM, Belarmino LC, Kido EA, Pandolfi V (2012) Overall picture of expressed Heat Shock Factors in Glycine max, Lotus japonicus, and Medicago truncatula. Genet Mol Biol 1:247–259
Song X, Liu G, Duan W, Liu T, Huang Z, Ren J, Li Y, Hou X (2014) Genome-wide identification, classification and expression analysis of the heat shock transcription factor family in Chinese cabbage. Mol Genet Genom 289:541–551
Stanke M, Morgenstern B (2005) AUGUSTUS: a web server for gene prediction in eukaryotes that allows user-defined constraints. Nucleic Acids Res 33:W465–W467
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729
Thudi M, Upadhyaya HD, Rathore A, Gaur PM, Krishnamurthy L, Roorkiwal M, Nayak SN, Chaturvedi SK, Basu PS, Gangarao NVPR, Fikre A, Kimurto P, Sharma PC, Sheshashayee MS, Tobita S, Kashiwagi J, Ito O, Killian A, Varshney RK (2014) Genetic dissection of drought and heat tolerance in chickpea through genome-wide and candidate gene-based association mapping approaches. PLoS One 9:e96758
Timothy LB, Mikael Bodén F, Buske A, Martin F, Charles EG, Luca C, Jingyuan R, Wilfred WL, William SN (2009) MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res 37:202–208
Varshney RK, Song C, Saxena RK, Azam S, Yu S, Sharpe AG, Cannon S, Baek J, Rosen BD, Tar’an B, Millan T, Zhang X, Ramsay LD, Iwata A, Wang Y, Nelson W, Farmer AD, Gaur PM, Soderlund C, Penmetsa RV, Xu C, Bharti AK, He W, Winter P, Zhao S, Hane JK, Carrasquilla-Garcia N, Condie JA, Upadhyaya HD, Luo M-C (2013) Draft genome sequence of chickpea (Cicer arietinum) provides a resource for trait improvement. Nat Biotechnol 31:240–246
Von Koskull-Döring P, Scharf KD, Nover L (2007) The diversity of plant heat stress transcription factors. Trends Plant Sci 12:452–457
Wang G, Zhang J, Moskophidis D, Mivechi NF (2003) Targeted disruption of the heat shock transcription factor (hsf)-2 gene results in increased embryonic lethality, neuronal defects, and reduced spermatogenesis. Genesis 36:48–61
Wang W, Vinocur B, Shoseyov O, Altman A (2004) Role of plant heat-shock proteins and molecular chaperones in the abiotic stress response. Trends Plant Sci 9:244–252
Wang F, Dong Q, Jiang H, Zhu S, Chen B, Xiang Y (2012a) Genome-wide analysis of the heat shock transcription factors in Populus trichocarpa and Medicago truncatula. Mol Biol Rep 39:1877–1886
Wang Y, Tang H, Debarry JD, Tan X, Li J, Wang X, Lee TH, Jin H, Marler B, Guo H, Kissinger JC, Paterson AH (2012b) MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res 40:e49
Wang J, Sun N, Deng T, Zhang L, Zuo K (2014) Genome-wide cloning, identification, classification and functional analysis of cotton heat shock transcription factors in cotton (Gossypium hirsutum). BMC Genom 15:961
Xin Q, Meng L, Leiting L, Hao Y, Juyou W, Shaoling Z (2015) Genome-wide identification and comparative analysis of the heat shock transcription factor family in Chinese white pear (Pyrus bretschneideri) and five other Rosaceae species. BMC Plant Biol 15:12
Xue GP, Sadat S, Drenth J, McIntyre CL (2014) The heat shock factor family from Triticum aestivum in response to heat and other major abiotic stresses and their role in the regulation of heat shock protein genes. J Exp Bot 65:539–557
Yang Z, Wang Y, Gao Y, Zhou Y, Zhang E, Hu Y, Yuan Y, Liang G, Xu C (2014) Adaptive evolution and divergent expression of heat stress transcription factors in grasses. BMC Evol Biol 14:147
Yoshida T, Ohama N, Nakajima J, Kidokoro S, Mizoi J, Nakashima K, Maruyama K, Kim J-M, Seki M, Todaka D, Osakabe Y, Sakuma Y, Schöffl F, Shinozaki K, Yamaguchi-Shinozaki K (2011) Arabidopsis HsfA1 transcription factors function as the main positive regulators in heat shock-responsive gene expression. Mol Genet Genom 286:321–332
Zafar SA, Hussain M, Raza M, Ahmed HGM-D, Rana IA, Sadia B, Atif RM (2016) Genome wide analysis of heat shock transcription factor (HSF) family in chickpea and its comparison with Arabidopsis. Plant Omics 9(2):136–141
Zhang L, Li Y, Xing D, Gao C (2009) Characterization of mitochondrial dynamics and subcellular localization of ROS reveal that HsfA2 alleviates oxidative damage caused by heat stress in Arabidopsis. J Exp Bot 60:2073–2091
Zhou S, Zhang P, Jing Z, Shi J (2013) Genome-wide identification and analysis of heat shock transcription factor family in cucumber (Cucumis sativus L.). Plant Omi J 6:449–455
Acknowledgements
The authors thank the Indian Council of Agricultural Research for financial assistance under the functional genomics component of Network Project on Transgenic Crops and the Emeritus Scientist Scheme to RS. PC acknowledges the SRF fellowship provided by Council of Scientific and Industrial Research (CSIR). JPTK and VS acknowledge the SRF fellowship provided by IARI. We also thank Dr. K.V. Prabhu and Dr. Rajinder Singh for providing the facilities to conduct the experiment in the National Phytotron Facility, IARI.
Author information
Authors and Affiliations
Corresponding author
Additional information
Prasanth Tej Kumar Jagannadham and Viswanathan Satheesh contributed equally to the work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Chidambaranathan, P., Jagannadham, P.T.K., Satheesh, V. et al. Genome-wide analysis identifies chickpea (Cicer arietinum) heat stress transcription factors (Hsfs) responsive to heat stress at the pod development stage. J Plant Res 131, 525–542 (2018). https://doi.org/10.1007/s10265-017-0948-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10265-017-0948-y