Abstract
Previous systematic reviews have found a higher sero-prevalence of EBV antibodies in SLE patients compared with controls. Because many studies have been published, there is a need to apply more precise systematic review methods. We examined the association between EBV and SLE patients by conducting a systematic review and meta-analysis of case–control studies that examined the prevalence of EBV antibodies and the DNA-positive rate. We searched the MEDLINE and EMBASE databases from 1966 to 2018 with no language restrictions. The Mantel–Haenszel odds ratios (OR) for EBV antibody sero-positivity were calculated, and meta-analyses were conducted. Quality assessment was performed using a modified version of the Newcastle–Ottawa scale, and 33 studies were included. Most studies found a higher sero-prevalence of VCA IgG and EA IgG in SLE patients compared with controls. Meta-analysis demonstrated a significantly higher OR for sero-positivity to VCA IgG and EA IgG for SLE cases (2.06 [95% confidence interval (CI) 1.30–3.26, p = 0.002] and 7.70, [95% CI 4.64–12.76, p < 0.001], respectively). The overall OR for the DNA-positive rate for SLE patients compared with controls was 3.86 (95% CI 1.52–9.83, p = 0.005). Other antibodies, i.e., VCA IgA/IgM, EBNA IgA, and EA IgA/IgM, also demonstrated a significant difference between SLE patients and controls. These findings support previous systematic reviews; however, publication bias cannot be excluded. The methodological conduct of studies could be improved, particularly when selecting controls and analyses of laboratory conduct.
Similar content being viewed by others
Background
Systemic lupus erythematosus is an autoimmune disease without clear pathogenesis. This disease is characterized by polyclonal B cell activation and altered T cell function with the presence of multiple autoantibodies and impaired cell-mediated immunity. It is believed that both genetic and environmental factors contribute to disease development [1]. Bacteria and virus infections are major important environmental factors that may be initiated and involved in the pathogenesis of SLE. The Epstein–Barr virus (EBV) is of particular interest. It has been reported that Epstein–Barr nuclear antigen-1 (EBNA-1) has a high degree of homology with some proteins that cause an autoimmune humoral response [2, 3]. This molecular mimicry may play an essential role in the induction of antibodies by EBV infection in SLE patients. Recently, Harley et al. [4] reported that nearly half of SLE risk loci are occupied by the EBNA-2 protein and many co-cluster with transcription factors, providing an essential new perspective on the mechanism of SLE pathogenesis.
The first positive association between EBV infection and SLE was found in 1971 [5]. Since then, many researchers have used various angles to investigate the possibility of this link. However, previous studies have failed to detect a consistent association. The first and only systematic review that updated on the association between SLE and sero-positivity for different EBV antibodies was that of Hanlon et al. [6]. These authors found a statistically significant higher sero-prevalence of viral capsid antigen (VCA) IgG but not EBNA1 in cases compared with controls. Meta-analyses for early antigen (EA)/D IgG and VCA IgA also significantly demonstrated higher ORs in cases compared with controls.
Many studies have since been published and may help in more fully estimating the association. In addition, some authors postulated that the increase in antibodies in SLE was brought about by generalized immune hyper-reactivity in lupus rather than by any specific property of the EBV. It was later thought that the best way to clarify this question would be at the DNA level [7,8,9,10]. Therefore, we undertook this meta-analysis with a broader, more comprehensive search strategy with no language restriction including both the sero-prevalence of antibodies and the EBV DNA load.
Methods
We conducted a systematic literature search of MEDLINE (source: PubMed, January 1, 1966 to June 20, 2018) and EMBASE (January 1, 1974 to June 20, 2018) using text and keywords in combination as MeSH terms and text words (Additional file: Table S1). We searched articles published without language restrictions and scrutinized the references from these studies to identify other relevant studies.
Study selection
To minimize differences between studies, we imposed the following methodological restrictions for the inclusion criteria: (1) The study had to be a cohort or case–control study of recruited patients with an SLE diagnosis and controls with no SLE diagnosis (healthy or unhealthy). (2) Patients could be from any age group, and studies that assayed the EBV DNA genome using peripheral blood mononuclear cells (PBMCs) and/or serum or antibodies (using any method) for any of the following EBV antigens: VCA, EA, EBNA-1, or 2. Exclusion criteria were as follows: (1) non-human studies. (2) Studies only measuring IgM antibodies. (3) Studies that reported serum antibodies without a specified antigen or an antigen without a specified antibody type.
Quality assessment
Because all of the included studies were non-randomized, we used the Newcastle–Ottawa scale (NOS) to access the quality of the studies [11], which was adapted to award two stars for blinded blood sample analysis, one star for conducting the analysis in a clinical laboratory (independent from the investigators), one star for specifying explicit laboratory cutoffs for sero-positivity, and one star for reporting the presence or absence of missing data.
Data analysis and statistical analysis
Two investigators (Li ZX and Zeng S) independently extracted data from eligible studies and reached a consensus for all items. For all studies, we extracted the following: the name of the first author, year of publication, country where the study was performed, number of patients, study population, gender category, selection criteria for control participants, methods for outcome assessment, and reported adjustments for potential confounders. The percent agreement between two authors on the review quality ranged from 85 to 100%.
The meta-analysis was performed by STATA 12.0 (Stata Corp, College Station, TX). We pooled estimated Mantel-Haenszel ORs from each study separately for each outcome using a random-effects meta-analysis. We evaluated the statistical heterogeneity of the ORs by calculating the Cochrane Q statistic (significance level: P ≤ 0.1) and the I2 statistic, applying the following interpretation for I2: < 50%, low heterogeneity, 50–75%, moderate heterogeneity, and > 75%, high heterogeneity [12]. The probability of publication bias was assessed by visual inspection of a funnel plot and Begg’s test [13, 14]. If the funnel plot and Begg’s test revealed asymmetry, we performed the Duval and Tweedie nonparametric “trim and fill” procedure to assess potential effects of publication bias [12]. This method considers the possibility of hypothetical “missing” studies that might exist, imputes their ORs, and recalculates a pooled OR that incorporates the hypothetical missing studies as though they exist. Subgroup analyses and meta-regression models were performed to investigate potential sources of between-study heterogeneity.
Results
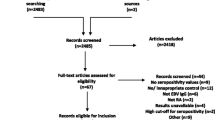
A total of 2000 references were retrieved using our initial search algorithm, and of these, 33 studies were ultimately included (Additional file: Figure S1). One study was in Turkish [15], one in Chinese [16], one in French [17], and the others were in English. All of the reports were case–control studies published between 1988 and 2018. There were two pediatric studies and 31 adult studies. The characteristics of the 33 eligible studies are listed in Table 1.
The features of the participants in the included studies are summarized in Table 1. There were 2814 cases and 4048 controls. The participants were almost all female with an average age of 37.5 years. The median sample sizes of the cases and controls were 85 and 123, respectively. Most of the studies specified using 1982 or 1997 American College of Rheumatology (ACR) criteria for SLE diagnoses (29 of 33 studies) for cases. The controls included healthy and non-healthy participants with the majority of the studies recruiting healthy controls. Only eight studies recruited samples from the general community. Most studies recruited hospital controls or did not state the source. There were four studies that recruited controls from patients’ relatives.
VCA (IgG, IgA, and IgM)
There were 20 studies that assayed VCA IgG sero-prevalence. We divided the study of Parks et al. into two separate studies, i.e., African-Americans and whites, making the total number of studies 21. This group found that SLE and the sero-prevalence of EBV antibodies were strongly associated in African-Americans and modestly associated in whites, reflecting significant interaction of race. These studies included a total of 1795 cases and 2635 controls. The mean sero-prevalence of VCA IgG in the cases and controls was 95.0 and 90.8%, respectively. The pooled OR from these studies was 2.06 (95% CI 1.30–3.26, p = 0.002) (Fig. 1a). The heterogeneity between studies was significant (Q = 40.77, p = 0.004, I2 = 50.9%). Visual inspection of a Begg’s funnel plot revealed asymmetry (p = 0.027). We further performed sensitivity analysis using the “trim and fill” procedure to assess the possible effects of publication bias, which involved using unpublished conservative studies to mirror the positive studies that caused plot asymmetry (Fig. 1b). The result demonstrated that no trimming was performed and the data were unchanged, suggesting that the association is not an artifact of unpublished negative studies.
There were six studies that measured VCA IgA, including Parks et al. These studies included a total of 431 cases and 431 controls. The overall percentage of those SLE positive with VCA IgA was higher than that for controls (45.0% and 21.3%, respectively). The pooled OR from these studies was 5.10 (95% CI 2.12–12.28, p < 0.001) (Additional file: Figure S2). The heterogeneity between the studies was also significant (Q = 18.91, p = 0.002, I2 = 73.6%). Because of the small number of included studies, a funnel plot was not created.
There were eight studies that measured VCA IgM, including Parks et al. These studies included a total of 665 cases and 648 controls. The overall percentage of those SLE positive with VCA IgM was higher than that for controls (21.1% and 8.3%, respectively). The pooled OR from these studies was 2.29 (95% CI 1.34–3.93, p = 0.003, I2 = 20.9%) (Additional file: Figure S3). A funnel plot was not created due to the small number of included studies.
EBNA (IgG, IgA)
We identified 19 studies that tested EBNA IgG sero-positivity, including data for 1572 SLE cases and 2270 controls. The mean sero-prevalence was slightly higher for the SLE group (90.2%) compared with that for controls (87.8%). The estimated pooled OR for EBNA IgG and SLE was 1.06 (95% CI 0.70–1.61, p = 0.787), suggesting no significant association between EBNA IgG and SLE (Fig. 2a). The heterogeneity between the studies was moderate (I2 = 52.9%). Visual inspection of a Begg’s funnel plot revealed symmetry (p = 0.484) (Fig. 2b).
Only three studies, including 209 SLE cases and 762 controls, demonstrated EBNA IgA sero-positivity. The estimated pooled OR for EBNA IgA and SLE was 10.40 (95% CI 6.51–16.62, p < 0.0001) (Additional file: Figure S4). The heterogeneity between the studies was low (I2 = 0.0%). A funnel plot was not created due to the small number of included studies.
EA (IgG, IgA, and IgM)
Twelve studies have reported the sero-positivity of EA IgG between SLE cases and controls. The mean sero-positivity for EA IgG was 60.6% (594/981) for SLE patients and 33.0% (409/1241) for controls. The estimated pooled OR for EA IgG sero-positivity and SLE was 7.70 (95% CI 4.64–12.76, p < 0.001) (Fig. 3a). Heterogeneity was moderate (Q = 29.9, p = 0.002, I2 = 63.2%). Visual inspection of a Begg’s funnel plot revealed a more symmetrical (p = 0.837) distribution of the studies, indicating that publication bias is less likely (Fig. 3b).
There were four studies that assayed the EA IgA sero-prevalence. The prevalence was extremely high in the SLE compared with control groups, 47.2% (91/193) and 4.3% (4/93), respectively. The estimated pooled OR was 16.06 (95% CI 5.99–43.04, p < 0.001) with low heterogeneity (Q = 1.09, p = 0.778, I2 = 0%) (Additional file: Figure S5).
There were three studies including 159 SLE cases and 71 controls that tested the EA IgM sero-prevalence. The estimated pooled OR was 4.21 (95% CI 2.11–8.40, p < 0.001), and heterogeneity was low (Q = 1.23, p = 0.540, I2 = 0%) (Additional file: Figure S6).
Ebv DNA
There were seven studies including 514 cases and 1086 controls that tested the EBV DNA-positive rate for the participants. The positive rate for DNA was 55.1% for the SLE group and 20.7% for the control group. Meta-analysis generated an overall OR of 3.864 (95% CI 1.518–9.830, p = 0.005) (Fig. 4). Heterogeneity was high with Q = 41.74 (p < 0.001) and I2 = 85.6%. Due to the small number of the studies, a funnel plot was not generated.
Subgroup analysis
To explore study heterogeneity, we performed subgroup analysis across a number of significant study characteristics. Age, sex, living place, diagnosis criteria for the cases, source of controls, and method for detecting antibodies were not a significant source of heterogeneity (Table 2). There was a trend that pediatric patients had higher ORs compared with adult patients, but there was no significant difference. According to some major studies, age was a significant confounding factor. We undertook an exploratory meta-regression analysis of the average age in each study. The results demonstrated that the average age significantly correlated with DNA positivity and SLE (p = 0.004) (Fig. 5).
The linear dose–response relationship between the DNA-positive rate and SLE with average age as the explanatory variable. The solid line represents point estimates of the association between EBV DNA positivity and SLE; the dashed lines are 95% CIs. Circles present the dose-specific OR estimates reported in each study. The area in each circle is proportional to the inverse variance of the OR. The vertical axis is on a log scale
Quality assessment
According to the modified NOS scale, the maximum score that could be achieved by a study was 12 stars. In our meta-analysis, the median score for all studies was five. The highest was rewarded by Parks et al. with nine stars. For selection criteria, only two studies did not specify a definition for the cases. However, only 6 of 33 recruited cases for consecutive or representative patients. Eight studies selected adequate controls from the community. For comparability criteria, 15 studies did not match cases and controls with confounders. Ten out of the remaining 18 studies matched for age and at least one additional factor. As for exposure, few studies reported the blinding of analyses or missing data. About half of the studies listed cutoffs for the assays.
To examine the influence of the quality of studies on ORs, we compared studies with higher NOS scores (equal to or above the median of the overall studies) to studies scoring below the median in a post hoc analysis. The ORs were higher for all EBV IgG and DNA outcomes in the higher scoring studies with the exception of EBNA IgG. However, there was no statically significant difference (Table 2).
Discussion
Our review has again found an association between EBV sero-positivity and SLE based on VCA antibody (IgG, IgA, IgM), EBNA IgA, and EA antibody (IgG, IgA, IgM) testing. We did not observe evidence of differences in the sero-prevalence of EBNA IgG, which is indicative of latent infection. This analysis shows a significant association between the EBV DNA-positive rate and SLE (OR: 3.86, 95% CI 1.52–9.83, p = 0.005). Furthermore, meta-regression demonstrates that the average age of the participants negatively correlated with the association between DNA positivity and SLE (p = 0.004). To our knowledge, this systemic review is the first attempt to combine such estimates of the association between SLE and EBV DNA positivity.
Hanlon et al. [6] included 25 studies in their meta-analysis, but four of the studies did not specify the antigen for the tested antibody. Therefore, only 21 of these studies were used for analysis. In our review, 12 additional studies were added for analysis. In addition to increasing the total number of cases, the average sample size also increased. As a matter of fact, our results even more precisely verify Hanlon’s findings.
There was considerable heterogeneity between studies, and we examined different factors that might have been influential. Although none of the subgroup analyses reached statistical significance. Slightly higher OR values were observed for the pediatric studies with a wide confidence interval (0.08–187.98) compared with adult studies. James et al. [8] found that EBV infection increases SLE risk by as much as 50-fold in children, while Tsai et al. [10] found no difference between SLE patients and healthy controls. This discrepancy may be due to the small number of pediatric studies (only two) or only chance.
We could not find any difference in the combined OR of the studies using more recent criteria (1997 ACR criteria for SLE) compared with older criteria (1982 ACR criteria for SLE) or failure to report criteria (which may reflect a lack of reporting). Studies matched for age, sex, or living place did not have different ORs for EBV IgG sero-positivity. Other potential vital confounders include ethnicity and socioeconomic status. We did not test the effects of controlling for these factors because few studies have specifically mentioned controlling for them. Parks et al. [29] reported a racial difference in the association between EBV IgA and SLE where African-Americans were more likely to have a history of EBV infection. Comparing the ORs of studies that used IFA to detect EBV antibodies vs. those that used ELISA revealed a similar result. However, Almohmeed et al. [45] demonstrated that using IFA to test VCA and EA IgG had twice the OR compared with ELISA for MS. Because there is a lack of detailed studies analyzing the two testing methods for SLE, we could not explain the discrepancy between these two autoimmune diseases. Other elements that may contribute to differences in estimates of ORs include the source and selection of patients and controls. Few studies have clearly described their methods for selecting cases (random or consecutive). Ideally, society would be the best source for controls. However, only eight studies have been deemed to have appropriate community controls [16, 18, 29, 33, 40,41,42, 44]. There were six studies that recruited bone marrow or blood donors [15, 17, 22, 28, 30, 31], which may be considered inappropriate community controls because such donors were selected with unhealthy risk behavior. There were four studies that recruited relatives of patients [8, 23, 35, 36], which are also inappropriate controls as relatives may share the same susceptible genetic factors as patients. Sixteen studies have recruited only seemingly healthy controls, and we observed that nine of these studies found no statistically significant higher EBV IgG sero-prevalence in cases compared with controls [19, 26,27,28,29, 34, 37, 41, 42].
The pooled OR for EBV DNA and SLE was 3.86, which is consistent with sero-antibody prevalence. Yu et al. [39] reported a high OR for an SLE patient that was significantly different from others because the controls had low positivity in serum samples. Although three of seven studies failed to find a significant difference in the DNA-positivity rate and SLE [9, 37, 44], and one found a high DNA load in SLE compared with controls [44]. The negative result may be due to chance or the small sizes of these studies. Yu et al. [39] reported that the EBV DNA-positive rate declines with age for controls but not for SLE patients. Although our subgroup analysis did not find a significant difference between age-matched and unmatched groups, meta-regression demonstrated that the OR for the DNA-positive rate declines with average age. This may be due to the fact that young SLE patients tend to be more infected by EBV, which is consistent with James et al. [8].
EBV subclinically infects a majority of individuals worldwide and generates multiple antibodies in the serum. The most common antibody is VCA IgG, which represents exposure to EBV and lasts lifelong. Our studies found that the overall prevalence of VCA IgG in SLE patients and controls was 95.0 and 90.8%, respectively, and the OR reached a significant difference. In addition, many studies have revealed a higher titer in SLE patients compared with controls [18, 23, 33,34,35]. Otherwise, the best way to specify the relationship between SLE and EBV infection is at the DNA level, which also had a higher OR in our study. A conclusion may be drawn that prior EBV infection is an essential factor for the development of SLE. Many studies had found that some humoral autoimmunity in SLE arises from molecular mimicry between EBNA-1 and lupus auto-antigens, which provide further evidence for suspecting an etiologic role for EBV in SLE. However, our study failed to find a higher prevalence of EBNA IgG in SLE patients, which is difficult to explain. Furthermore, a prospective study with a large sample size should be considered to solve this enigma. Antibodies directed against EA/D are generally known as an indication of lytic replication. The sero-prevalence of these antibodies is significantly higher for SLE patients, particularly for IgA, compared with controls, and the ORs also achieved statistical significance. The majority of the included studies have been consistent on this issue. Draborg et al. [33] thought that a possible mechanism for this phenomenon as a specific intrinsic defect in the immune systems of SLE patients is independent of immunosuppressive medication therapy.
The strengths of this meta-analysis include its comprehensive search strategy and absence of language restrictions; however, limitations must also be considered. First, the quality of individual studies was not always ideal, as demonstrated by a general lack of information on the recruitment of consecutive patients and selection controls for all studies. Second, there is heterogeneity in the ORs across studies, and subgroup analysis did not reveal the source. This heterogeneity may be derived from other confounders, such as ethnicity and laboratory measurements. Third, some of the funnel plot analyses showed asymmetry that suggested the possibility of publication bias. The trim and fill sensitivity analysis did not modify the general results, suggesting that association is not an artifact of unpublished negative studies. Nevertheless, that possibility is not entirely excluded by this method. Fourth, similar to all meta-analyses, our study has the limitation of being a retrospective analysis; thus, further prospective cohort studies are warranted to confirm these findings.
In conclusion, our review supports the hypothesis that prior EBV infection is an essential factor for the development of SLE as indicated by a higher positivity of VCA IgG and EBV DNA. These findings also suggest abnormal (EA, IgA, EBV DNA) humoral immune responses to EBV in the context of SLE. However, the studies included in our meta-analysis are heterogeneous and have small sample sizes. At the same time, many studies did not match for age and gender, only a few matched for race and other confounding factors, and descriptions of recruitment and laboratory testing were not specified in most of the papers. Moreover, the role of publication bias could not be excluded. Large prospective studies are needed to determine the relationship between SLE and infection before we could draw a causal relationship between the two.
Abbreviations
- EBV:
-
Epstein–Barr virus
- SLE:
-
Systemic lupus erythematosus
- VCA:
-
Viral capsid antigen
- EA:
-
Early antigen
- EBNA:
-
Epstein–Barr nuclear antigen
- OR:
-
Odds ratio
- NOS:
-
Newcastle–Ottawa scale
- ACR:
-
American College of Rheumatology
- PBMCs:
-
Blood mononuclear cells
- ELISA:
-
Enzyme-linked immuno sorbent assay
- PCR:
-
Polymerase chain reaction
- IFA:
-
Indirect immunofluorescence assay
References
Munz C, Lunemann JD, Getts MT, Miller SD. Antiviral immune responses: triggers of or triggered by autoimmunity? Nat Rev Immunol. 2009;9(4):246–58.
James JA, Scofield RH, Harley JB. Lupus humoral autoimmunity after short peptide immunization. Ann N Y Acad Sci. 1997;815:124–7.
McClain MT, Heinlen LD, Dennis GJ, Roebuck J, Harley JB, James JA. Early events in lupus humoral autoimmunity suggest initiation through molecular mimicry. Nat Med. 2005;11(1):85–9.
Harley JB, Chen X, Pujato M, et al. Transcription factors operate across disease loci, with EBNA2 implicated in autoimmunity. Nat Genet. 2018;50(5):699–707.
Evans AS, Rothfield NF, Niederman JC. Raised antibody titres to EB virus in systemic lupus erythematosus. Lancet. 1971;1(7691):167–8.
Hanlon P, Avenell A, Aucott L, Vickers MA. Systematic review and meta-analysis of the sero-epidemiological association between Epstein-Barr virus and systemic lupus erythematosus. Arthritis Res Ther. 2014;16(1):R3.
Incaprera M, Rindi L, Bazzichi A, Garzelli C. Potential role of the Epstein-Barr virus in systemic lupus erythematosus autoimmunity. Clin Exp Rheumatol. 1998;16(3):289–94.
James JA, Kaufman KM, Farris AD, Taylor-Albert E, Lehman TJ, Harley JB. An increased prevalence of Epstein-Barr virus infection in young patients suggests a possible etiology for systemic lupus erythematosus. J Clin Invest. 1997;100(12):3019–26.
Lau CS, Yuen KY, Chan KH, Wong RW. Lack of evidence of active lytic replication of Epstein-Barr and cytomegaloviruses in patients with systemic lupus erythematosus. Chin Med J (Engl). 1998;111(7):660–5.
Tsai YT, Chiang BL, Kao YF, Hsieh KH. Detection of Epstein-Barr virus and cytomegalovirus genome in white blood cells from patients with juvenile rheumatoid arthritis and childhood systemic lupus erythematosus. Int Arch Allergy Immunol. 1995;106(3):235–40.
Wells GA, Shea B, O’Connell D, et al. The Newcastle -Ottawa Scale (NOS) for assessing the quality if nonrandomized studies in meta-analyses. http://www.ohrica/programs/clinical_epidemiology/oxfordasp 2011.
Higgins JPT, Thompson SG. Quantifying heterogeneity in a meta-analysis. Stat Med. 2002;21(11):1539–58.
Begg CB, Mazumdar M. Operating characteristics of a rank correlation test for publication bias. Biometrics. 1994;50(4):1088–101.
Egger M, Davey Smith G, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315(7109):629–34.
Us T, Cetin E, Kasifoglu N, Kasifoglu T, Akgun Y. Investigation of Epstein-Barr virus and herpes simplex virus markers by serological and molecular methods in patients with rheumatoid arthritis and systemic lupus erythematosus. Mikrobiyol Bul. 2011;45(4):677–83.
Zhang X, Li B, Liu Y, Jiang M. Clinical study on antibodies against EBV in sera of patients with rheumatoid arthritis. Zhongguo Yi Xue Ke Xue Yuan Xue Bao. 1999;21(1):8–12.
Tazi I, Fehri S, Elghrari K, Ouazzani T, Benchemsi N. Systemic lupus erythematosus and Epstein-Barr virus. East Mediterr Health J. 2009;15(3):701–8.
Berkun Y, Zandman-Goddard G, Barzilai O, et al. Infectious antibodies in systemic lupus erythematosus patients. Lupus. 2009;18(13):1129–35.
Chen CJ, Lin KH, Lin SC, et al. High prevalence of immunoglobulin A antibody against Epstein-Barr virus capsid antigen in adult patients with lupus with disease flare: case control studies. J Rheumatol. 2005;32(1):44–7.
Chen DY, Chen YM, Lan JL, et al. Polymyositis/dermatomyositis and nasopharyngeal carcinoma: the Epstein-Barr virus connection? J Clin Virol. 2010;49(4):290–5.
Esen BA, Yilmaz G, Uzun S, et al. Serologic response to Epstein-Barr virus antigens in patients with systemic lupus erythematosus: a controlled study. Rheumatol Int. 2012;32(1):79–83.
Huggins ML, Todd I, Powell RJ. Reactivation of Epstein-Barr virus in patients with systemic lupus erythematosus. Rheumatol Int. 2005;25(3):183–7.
James JA, Neas BR, Moser KL, et al. Systemic lupus erythematosus in adults is associated with previous Epstein-Barr virus exposure. Arthritis Rheum. 2001;44(5):1122–6.
Kitagawa H, Iho S, Yokochi T, Hoshino T. Detection of antibodies to the Epstein-Barr virus nuclear antigens in the sera from patients with systemic lupus erythematosus. Immunol Lett. 1988;17(3):249–52.
Lu JJ, Chen DY, Hsieh CW, Lan JL, Lin FJ, Lin SH. Association of Epstein-Barr virus infection with systemic lupus erythematosus in Taiwan. Lupus. 2007;16(3):168–75.
Marchini B, Dolcher MP, Sabbatini A, Klein G, Migliorini P. Immune response to different sequences of the EBNA I molecule in Epstein-Barr virus-related disorders and in autoimmune diseases. J Autoimmun. 1994;7(2):179–91.
Newkirk MM, Shiroky JB, Johnson N, et al. Rheumatic disease patients, prone to Sjogren’s syndrome and/or lymphoma, mount an antibody response to BHRF1, the Epstein-Barr viral homologue of BCL-2. Br J Rheumatol. 1996;35(11):1075–81.
Ngou J, Segondy M. Immunoblotting reactivity of sera from patients with autoimmune connective tissue diseases against Epstein-Barr nuclear antigen (EBNA) polypeptides. Serodiagn Immunother Infect Dis. 1996;8(2):105–8.
Parks CG, Cooper GS, Hudson LL, et al. Association of Epstein-Barr virus with systemic lupus erythematosus: effect modification by race, age, and cytotoxic T lymphocyte-associated antigen 4 genotype. Arthritis Rheum. 2005;52(4):1148–59.
Stratta P, Canavese C, Ciccone G, et al. Correlation between cytomegalovirus infection and Raynaud’s phenomenon in lupus nephritis. Nephron. 1999;82(2):145–54.
Westgeest AA, van Loon AM, van der Logt JT, van de Putte LB, Boerbooms AM. Antiperinuclear factor, a rheumatoid arthritis specific autoantibody: its relation to Epstein-Barr virus. J Rheumatol. 1989;16(5):626–30.
Yokochi T, Yanagawa A, Kimura Y, Mizushima Y. High titer of antibody to the Epstein-Barr virus membrane antigen in sera from patients with rheumatoid arthritis and systemic lupus erythematosus. J Rheumatol. 1989;16(8):1029–32.
Draborg AH, Jorgensen JM, Muller H, et al. Epstein-Barr virus early antigen diffuse (EBV-EA/D)-directed immunoglobulin A antibodies in systemic lupus erythematosus patients. Scand J Rheumatol. 2012;41(4):280–9.
Chougule D, Nadkar M, Rajadhyaksha A, et al. Association of clinical and serological parameters of systemic lupus erythematosus patients with Epstein-Barr virus antibody profile. J Med Virol. 2018;90(3):559–63.
Csuka D, Simon D, Hobor R, et al. Serum concentration of immunoglobulin G-type antibodies against the whole Epstein-Barr nuclear antigen 1 and its aa35-58 or aa398-404 fragments in the sera of patients with systemic lupus erythematosus and multiple sclerosis. Clin Exp Immunol. 2013;171(3):255–62.
Vista ES, Weisman MH, Ishimori ML, et al. Strong viral associations with SLE among Filipinos. Lupus Sci Med. 2017;4(1):e000214.
Broccolo F, Drago F, Cassina G, et al. Selective reactivation of human herpesvirus 6 in patients with autoimmune connective tissue diseases. J Med Virol. 2013;85(11):1925–34.
Han L, Zhang Y, Wang Q, et al. Epstein-Barr virus infection and type I interferon signature in patients with systemic lupus erythematosus. Lupus. 2018. https://doi.org/10.1177/0961203317753069.
Yu SF, Wu HC, Tsai WC, et al. Detecting Epstein-Barr virus DNA from peripheral blood mononuclear cells in adult patients with systemic lupus erythematosus in Taiwan. Med Microbiol Immunol. 2005;194(3):115–20.
Rasmussen NS, Draborg AH, Nielsen CT, Jacobsen S, Houen G. Antibodies to early EBV, CMV, and HHV6 antigens in systemic lupus erythematosus patients. Scand J Rheumatol. 2015;44(2):143–9.
Draborg AH, Jacobsen S, Westergaard M, et al. Reduced response to Epstein-Barr virus antigens by T-cells in systemic lupus erythematosus patients. Lupus Sci Med. 2014;1(1):e000015.
Draborg AH, Sandhu N, Larsen N, Lisander Larsen J, Jacobsen S, Houen G. Impaired cytokine responses to Epstein-Barr virus antigens in systemic lupus Erythematosus patients. J Immunol Res. 2016;2016:6473204.
Larsen M, Sauce D, Deback C, et al. Exhausted cytotoxic control of Epstein-Barr virus in human lupus. PLoS Pathog. 2011;7(10):e1002328.
Moon UY, Park SJ, Oh ST, et al. Patients with systemic lupus erythematosus have abnormally elevated Epstein-Barr virus load in blood. Arthritis Res Ther. 2004;6(4):R295–302.
Almohmeed YH, Avenell A, Aucott L, Vickers MA. Systematic review and meta-analysis of the sero-epidemiological association between Epstein Barr virus and multiple sclerosis. PLoS ONE. 2013;8(4):e61110.
Funding
It provided financial support for submitting the paper for publication.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Ethical approval
All analyses were based on previous published studies; thus, no ethical approval is required.
Informed consent
All analyses were based on previous published studies; thus, no informed consent is required.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Li, ZX., Zeng, S., Wu, HX. et al. The risk of systemic lupus erythematosus associated with Epstein–Barr virus infection: a systematic review and meta-analysis. Clin Exp Med 19, 23–36 (2019). https://doi.org/10.1007/s10238-018-0535-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10238-018-0535-0