Abstract
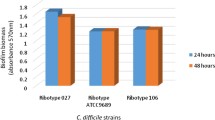
Clostridium difficile is the cause of the nosocomial C. difficile infection (CDI). The conventional antibiotics used in CDI therapy are often unsuccessful, and recurrent infections may occur. Biofilm formation by C. difficile is associated with chronic or recurrent infections; biofilms may contribute to virulence and impaired antimicrobial efficacy. Manuka honey, derived from the Manuka tree (Leptospermum scoparium), is known to exhibit antimicrobial properties that are associated with its significant content of methylglyoxal, a natural antibiotic. The aim of the present study was to determine the antimicrobial effect of Manuka honey on clinical C. difficile strains belonging to four prominent polymerase chain reaction (PCR) ribotypes (RTs) (RT017, RT023, RT027 and RT046) and on their biofilm formation in vitro. Minimal inhibitory and bactericidal concentrations (MICs and MBCs, respectively) were determined using the broth dilution method. The biomass of the biofilm and the clearance of C. difficile biofilms by Manuka honey were determined using the crystal violet staining method. The MIC and MBC of Manuka honey for C. difficile strains were equal at 6.25% (v/v). PCR RT027 strains produced more biofilm in vitro than the other examined strains. Manuka honey effectively inhibited biofilm formation by C. difficile strains of different PCR RTs.


Similar content being viewed by others
References
Rupnik M, Wilcox MH, Gerding DN (2009) Clostridium difficile infection: new developments in epidemiology and pathogenesis. Nat Rev Microbiol 7:526–536. doi:10.1038/nrmicro2164
McFarland LV, Surawicz CM, Stamm WE (1990) Risk factors for Clostridium difficile carriage and C. difficile-associated diarrhea in a cohort of hospitalized patients. J Infect Dis 162:678–684
Davies KA, Ashwin H, Longshaw CM, Burns DA, Davis GL, Wilcox MH; EUCLID study group (2016) Diversity of Clostridium difficile PCR ribotypes in Europe: results from the European, multicentre, prospective, biannual, point-prevalence study of Clostridium difficile infection in hospitalised patients with diarrhoea (EUCLID), 2012 and 2013. Euro Surveill 21(29). doi:10.2807/1560-7917.ES.2016.21.29.30294
Pituch H, Obuch-Woszczatyński P, Lachowicz D, Wultańska D, Karpiński P, Młynarczyk G, van Dorp SM, Kuijper EJ; Polish Clostridium difficile Study Group (2015) Hospital-based Clostridium difficile infection surveillance reveals high proportions of PCR ribotypes 027 and 176 in different areas of Poland, 2011 to 2013. Euro Surveill 20(38). doi: 10.2807/1560-7917.ES.2015.20.38.30025
Bartlett JG (2006) Narrative review: the new epidemic of Clostridium difficile-associated enteric disease. Ann Intern Med 145:758–764
Vedantam G, Clark A, Chu M, McQuade R, Mallozzi M, Viswanathan VK (2012) Clostridium difficile infection: toxins and non-toxin virulence factors, and their contributions to disease establishment and host response. Gut Microbes 3:121–134. doi:10.4161/gmic.19399
Tasteyre A, Barc MC, Karjalainen T, Dodson P, Hyde S, Bourlioux P, Borriello P (2000) A Clostridium difficile gene encoding flagellin. Microbiology 146:957–966
Chapetón Montes D, Candela T, Collignon A, Janoir C (2011) Localization of the Clostridium difficile cysteine protease Cwp84 and insights into its maturation process. J Bacteriol 193:5314–5321. doi:10.1128/JB.00326-11
Ðapa T, Leuzzi R, Ng YK, Baban ST, Adamo R, Kuehne SA, Scarselli M, Minton NP, Serruto D, Unnikrishnan M (2013) Multiple factors modulate biofilm formation by the anaerobic pathogen Clostridium difficile. J Bacteriol 195:545–555. doi:10.1128/JB.01980-12
Malone M, Bjarnsholt T, McBain AJ, James GA, Stoodley P, Leaper D, Tachi M, Schultz G, Swanson T, Wolcott RD (2017) The prevalence of biofilms in chronic wounds: a systematic review and meta-analysis of published data. J Wound Care 26:20–25. doi:10.12968/jowc.2017.26.1.20
Donlan RM (2001) Biofilms and device-associated infections. Emerg Infect Dis 7:277–281
Crowther GS, Chilton CH, Todhunter SL, Nicholson S, Freeman J, Baines SD, Wilcox MH (2014) Comparison of planktonic and biofilm-associated communities of Clostridium difficile and indigenous gut microbiota in a triple-stage chemostat gut model. J Antimicrob Chemother 69:2137–2147. doi:10.1093/jac/dku116
Mah TF, O’Toole GA (2001) Mechanisms of biofilm resistance to antimicrobial agents. Trends Microbiol 9:34–39
Mathur H, Rea MC, Cotter PD, Hill C, Ross RP (2016) The efficacy of thuricin CD, tigecycline, vancomycin, teicoplanin, rifampicin and nitazoxanide, independently and in paired combinations against Clostridium difficile biofilms and planktonic cells. Gut Pathog 8:20. doi:10.1186/s13099-016-0102-8
Hammond EN, Donkor ES, Brown CA (2014) Biofilm formation of Clostridium difficile and susceptibility to Manuka honey. BMC Complement Altern Med 14:329. doi:10.1186/1472-6882-14-329
Stubbs SLJ, Brazier JS, O’Neill GL, Duerden BI (1999) PCR targeted to the 16S–23S rRNA gene intergenic spacer region of Clostridium difficile and construction of a library consisting of 116 different PCR ribotypes. J Clin Microbiol 37:461–463
Stepanovic S, Vukovic D, Dakic I, Savic B, Svabic-Vlahovic M (2000) A modified microtiter-plate test for quantification of staphylococcal biofilm formation. J Microbiol Methods 40:175–179
Maddocks SE, Lopez MS, Rowlands RS, Cooper RA (2012) Manuka honey inhibits the development of Streptococcus pyogenes biofilms and causes reduced expression of two fibronectin binding proteins. Microbiology 158:781–790. doi:10.1099/mic.0.053959-0
Anthimidou E, Mossialos D (2013) Antibacterial activity of Greek and Cypriot honeys against Staphylococcus aureus and Pseudomonas aeruginosa in comparison to manuka honey. J Med Food 16:42–47. doi:10.1089/jmf.2012.0042
Cooper RA, Halas E, Molan PC (2002) The efficacy of honey in inhibiting strains of Pseudomonas aeruginosa from infected burns. J Burn Care Rehabil 23:366–370
Hammond EN, Donkor ES (2013) Antibacterial effect of Manuka honey on Clostridium difficile. BMC Res Notes 6:188. doi:10.1186/1756-0500-6-188
Acknowledgements
We thank all the microbiologists who provided isolates of C. difficile to the Anaerobe Laboratory (AL), Department of Medical Microbiology, Medical University of Warsaw.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Isolates were collected as part of routine hospitals surveillance. Ethical approval and informed consent were not required.
Funding
This research received no specific grant.
Conflict of interest
None to declare except that Alex van Belkum is an employee of bioMérieux Inc.
Additional information
Data availability: The data have been deposited in the Figshare Digital Repository. doi:10.6084/m9.figshare.4781224.v3
Rights and permissions
About this article
Cite this article
Piotrowski, M., Karpiński, P., Pituch, H. et al. Antimicrobial effects of Manuka honey on in vitro biofilm formation by Clostridium difficile . Eur J Clin Microbiol Infect Dis 36, 1661–1664 (2017). https://doi.org/10.1007/s10096-017-2980-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10096-017-2980-1