Abstract
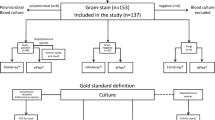
The use of molecular assays to rapidly identify pathogens and resistance genes directly from positive blood cultures (BCs) contribute to shortening the time required for the diagnosis of bloodstream infections. In this work, loop-mediated isothermal amplification (LAMP) assays have been examined for their potential use in BC diagnosis. Three different assays were applied. The commercially available eazyplex® MRSA test detects Staphylococcus aureus, S. epidermidis, mecA, and mecC. Two in-house assays [Gram-positive (GP) and Gram-negative (GN)] have been developed for the detection of streptococci, enterococci, vanA, vanB, Pseudomonas spp., Enterobacteriaceae, and the bla CTX-M family. A total of 370 positive BCs were analyzed. LAMP test results were obtained within 30 min, including sample preparation. Amplification was measured by real-time fluorescence detection. The threshold time for fluorescence intensity values ranged from 6.25 to 13.75 min. The specificity and sensitivity of the assays varied depending on the target. Overall, from 87.7% of BCs, true-positive results were obtained, compared to routine standard diagnosis. Twenty-one tests were true-negative because of the lack of an appropriate target (5.7%). The concordance of positive test results for resistance genes with subsequent antibiotic susceptibility testing was 100%. From 15 BC bottles with mixed cultures, eazyplex® assays produced correct results in 73% of the cases. This study shows that LAMP assays are fast and cost-saving tools for rapid BC testing in order to expedite the diagnostic report and improve the antibiotic stewardship for sepsis patients.
Similar content being viewed by others
References
Zilberberg MD, Shorr AF, Micek ST, Vazquez-Guillamet C, Kollef MH (2014) Multi-drug resistance, inappropriate initial antibiotic therapy and mortality in Gram-negative severe sepsis and septic shock: a retrospective cohort study. Crit Care 18:596
Fitting C, Parlato M, Adib-Conquy M, Memain N, Philippart F, Misset B, Monchi M, Cavaillon JM, Adrie C (2012) DNAemia detection by multiplex PCR and biomarkers for infection in systemic inflammatory response syndrome patients. PLoS One 7, e38916
Vincent JL, Brealey D, Libert N, Abidi NE, O’Dwyer M, Zacharowski K, Mikaszewska-Sokolewicz M, Schrenzel J, Simon F, Wilks M, Picard-Maureau M, Chalfin DB, Ecker DJ, Sampath R, Singer M; Rapid Diagnosis of Infections in the Critically Ill Team (2015) Rapid diagnosis of infection in the critically ill, a multicenter study of molecular detection in bloodstream infections, pneumonia, and sterile site infections. Crit Care Med 43:2283–2291
Rödel J, Karrasch M, Edel B, Stoll S, Bohnert J, Löffler B, Saupe A, Pfister W (2016) Antibiotic treatment algorithm development based on a microarray nucleic acid assay for rapid bacterial identification and resistance determination from positive blood cultures. Diagn Microbiol Infect Dis 84:252–257
Salimnia H, Fairfax MR, Lephart PR, Schreckenberger P, DesJarlais SM, Johnson JK, Robinson G, Carroll KC, Greer A, Morgan M, Chan R, Loeffelholz M, Valencia-Shelton F, Jenkins S, Schuetz AN, Daly JA, Barney T, Hemmert A, Kanack KJ (2016) Evaluation of the FilmArray blood culture identification panel: results of a multicenter controlled trial. J Clin Microbiol 54:687–698
Rand KH, Delano JP (2014) Direct identification of bacteria in positive blood cultures: comparison of two rapid methods, FilmArray and mass spectrometry. Diagn Microbiol Infect Dis 79:293–297
Alby K, Daniels LM, Weber DJ, Miller MB (2013) Development of a treatment algorithm for streptococci and enterococci from positive blood cultures identified with the Verigene Gram-positive blood culture assay. J Clin Microbiol 51:3869–3871
Ward C, Stocker K, Begum J, Wade P, Ebrahimsa U, Goldenberg SD (2015) Performance evaluation of the Verigene® (Nanosphere) and FilmArray® (BioFire®) molecular assays for identification of causative organisms in bacterial bloodstream infections. Eur J Clin Microbiol Infect Dis 34:487–496. doi:10.1007/s10096-014-2252-2
Southern TR, VanSchooneveld TC, Bannister DL, Brown TL, Crismon AS, Buss SN, Iwen PC, Fey PD (2015) Implementation and performance of the BioFire FilmArray® blood culture identification panel with antimicrobial treatment recommendations for bloodstream infections at a midwestern academic tertiary hospital. Diagn Microbiol Infect Dis 81:96–101
Notomi T, Okayama H, Masubuchi H, Yonekawa T, Watanabe K, Amino N, Hase T (2000) Loop-mediated isothermal amplification of DNA. Nucleic Acids Res 28, e63
Goldmeyer J, Li H, McCormac M, Cook S, Stratton C, Lemieux B, Kong H, Tang W, Tang YW (2008) Identification of Staphylococcus aureus and determination of methicillin resistance directly from positive blood cultures by isothermal amplification and a disposable detection device. J Clin Microbiol 46:1534–1536
Francois P, Tangomo M, Hibbs J, Bonetti EJ, Boehme CC, Notomi T, Perkins MD, Schrenzel J (2011) Robustness of a loop-mediated isothermal amplification reaction for diagnostic applications. FEMS Immunol Med Microbiol 62:41–48
García-Fernández S, Morosini MI, Marco F, Gijón D, Vergara A, Vila J, Ruiz-Garbajosa P, Cantón R (2015) Evaluation of the eazyplex® SuperBug CRE system for rapid detection of carbapenemases and ESBLs in clinical Enterobacteriaceae isolates recovered at two Spanish hospitals. J Antimicrob Chemother 70:1047–1050
Findlay J, Hopkins KL, Meunier D, Woodford N (2015) Evaluation of three commercial assays for rapid detection of genes encoding clinically relevant carbapenemases in cultured bacteria. J Antimicrob Chemother 70:1338–1342
Hinić V, Ziegler J, Straub C, Goldenberger D, Frei R (2015) Extended-spectrum β-lactamase (ESBL) detection directly from urine samples with the rapid isothermal amplification-based eazyplex® SuperBug CRE assay: proof of concept. J Microbiol Methods 119:203–205
Zboromyrska Y, Vergara A, Cosgaya C, Verger G, Mosqueda N, Almela M, Pitart C, Roca I, Marco F, Vila J (2015) Rapid detection of β-lactamases directly from positive blood cultures using a loop-mediated isothermal amplification (LAMP)-based assay. Int J Antimicrob Agents 46:355–356
Box MJ, Sullivan EL, Ortwine KN, Parmenter MA, Quigley MM, Aguilar-Higgins LM, MacIntosh CL, Goerke KF, Lim RA (2015) Outcomes of rapid identification for Gram-positive bacteremia in combination with antibiotic stewardship at a community-based hospital system. Pharmacotherapy 35:269–276
Roshdy DG, Tran A, LeCroy N, Zeng D, Ou FS, Daniels LM, Weber DJ, Alby K, Miller MB (2015) Impact of a rapid microarray-based assay for identification of positive blood cultures for treatment optimization for patients with streptococcal and enterococcal bacteremia. J Clin Microbiol 53:1411–1414
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Ethical approval
The study protocol for the evaluation of LAMP eazyplex® assays for clinical BCs was reviewed and approved by the ethics committee of the Friedrich Schiller University of Jena (4400-04/15).
Funding information
This study was supported by internal funding from the Jena University Hospital and by a grant from the German Federal Ministry of Education and Research (BMBF, 13N13890).
Conflict of interest
J. Rödel received financial support from Amplex BioSystems to attend the 67th Annual Meeting of the German Society for Hygiene and Microbiology in 2015. No funding was received from the manufacturer of eazyplex® tests for the purpose of the study.
Rights and permissions
About this article
Cite this article
Rödel, J., Bohnert, J.A., Stoll, S. et al. Evaluation of loop-mediated isothermal amplification for the rapid identification of bacteria and resistance determinants in positive blood cultures. Eur J Clin Microbiol Infect Dis 36, 1033–1040 (2017). https://doi.org/10.1007/s10096-016-2888-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10096-016-2888-1