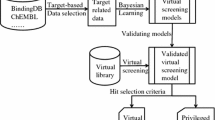
Abstract
The demand for new therapies has encouraged the development of faster and cheaper methods of drug design. Considering the number of potential biological targets for new drugs, the docking-based virtual screening (DBVS) approach has occupied a prominent role among modern strategies for identifying new bioactive substances. Some tools have been developed to validate docking methodologies and identify false positives, such as the receiver operating characteristic (ROC) curve. In this context, a database with 31 molecular targets called the Our Own Molecular Targets Data Bank (OOMT) was validated using the root-mean-square deviation (RMSD) and the area under the ROC curve (AUC) with two different docking methodologies: AutoDock Vina and DOCK 6. Sixteen molecular targets showed AUC values of >0.8, and those targets were selected for molecular docking studies. The drug-likeness properties were then determined for 473 Brazilian natural compounds that were obtained from the ZINC database. Ninety-six compounds showed similar drug-likeness property values to the marked drugs (positive values). These compounds were submitted to DBVS for 16 molecular targets. Our results showed that AutoDock Vina was more appropriate than DOCK 6 for performing DBVS experiments. Furthermore, this work suggests that three compounds—ZINC13513540, ZINC06041137, and ZINC1342926—are inhibitors of the three molecular targets 1AGW, 2ZOQ, and 3EYG, respectively, which are associated with cancer. Finally, since ZINC and the PDB were solely created to store biomolecule structures, their utilization requires the application of filters to improve the first steps of the drug development process.

Evaluation of docking methods used for virtual screening


Similar content being viewed by others
References
Khanna I (2012) Drug discovery in pharmaceutical industry: productivity challenges and trends. Drug Discov Today 17:1088–1102. doi:10.1016/j.drudis.2012.05.007
Geromichalos GD, Alifieris CE, Geromichalou EG, Trafalis DT (2016) Overview on the current status of virtual high-throughput screening and combinatorial chemistry approaches in multi-target anticancer drug discovery; part I. J Buon 21:764–779
Cheng T, Li Q, Zhou Z et al (2012) Structure-based virtual screening for drug discovery: a problem-centric review. AAPS J 14:133–41. doi:10.1208/s12248-012-9322-0
Lauro G, Romano A, Riccio R, Bifulco G (2011) Inverse virtual screening of antitumor targets: pilot study on a small database of natural bioactive compounds. J Nat Prod 74:1401–1407. doi:10.1021/np100935s
Glaab E (2016) Building a virtual ligand screening pipeline using free software: a survey. Brief Bioinform 17:352–366. doi:10.1093/bib/bbv037
Bursulaya BD, Bursulaya BD, Totrov M et al (2004) Comparative study of several algorithms for flexible ligand docking. J Comput Aided Mol Des 17:755–763
Ding Y, Fang Y, Moreno J et al (2016) Assessing the similarity of ligand binding conformations with the contact mode score. Comput Biol Chem 64:403–413. doi:10.1016/j.compbiolchem.2016.08.007
Triballeau N, Acher F, Brabet I et al (2005) Virtual screening workflow development guided by the “receiver operating characteristic” curve approach. Application to high-throughput docking on metabotropic glutamate receptor subtype 4. J Med Chem 48:2534–2547. doi:10.1021/jm049092j
Zhao W, Hevener KE, White SW et al (2009) A statistical framework to evaluate virtual screening. BMC Bioinformatics 10:225. doi:10.1186/1471-2105-10-225
Lätti S, Niinivehmas S, Pentikäinen OT (2016) Rocker: open source, easy-to-use tool for AUC and enrichment calculations and ROC visualization. J Cheminform 8:45. doi:10.1186/s13321-016-0158-y
Westermaier Y, Barril X, Scapozza L (2015) Virtual screening: an in silico tool for interlacing the chemical universe with the proteome. Methods 71:44–57. doi:10.1016/j.ymeth.2014.08.001
Li JW-H, Vederas JC (2009) Drug discovery and natural products: end of an era or an endless frontier? Science 325:161–165. doi: 10.1126/science.1168243
Trott O, Olson AJ (2009) AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem 31:455–461. doi:10.1002/jcc.21334.
Carregal AP, Comar M, Alves SN et al (2012) Inverse virtual screening studies of selected natural compounds from Cerrado. Int J Quantum Chem 112:3333–3340. doi:10.1002/qua.24205
Carregal AP, Comar M Jr, Taranto AG (2013) Our Own Molecular Targets Data Bank (OOMT). Biochem Biotechnol Rep 2:14–16
Rose PW, Prlić A, Bi C et al (2015) The RCSB Protein Data Bank: views of structural biology for basic and applied research and education. Nucleic Acids Res 43:D345–D356. doi:10.1093/nar/gku1214
Irwin JJ, Sterling T, Mysinger MM et al (2012) ZINC: a free tool to discover chemistry for biology. J Chem Inf Model 52:1757–1768. doi:10.1021/ci3001277
Walters WP, Murcko MA (2002) Prediction of “drug-likeness”. Adv Drug Deliv Rev 54:255–271. doi:10.1016/S0169-409X(02)00003-0
Gaulton A, Bellis LJ, Bento AP et al (2012) ChEMBL: a large-scale bioactivity database for drug discovery. Nucleic Acids Res 40:1100–1107. doi:10.1093/nar/gkr777
Bento AP, Gaulton A, Hersey A et al (2014) The ChEMBL bioactivity database: an update. Nucleic Acids Res 42:D1083–D1090. doi:10.1093/nar/gkt1031
Huang N, Shoichet BK, Irwin JJ (2006) Benchmarking sets for molecular docking. J Med Chem 49:6789–6801. doi:10.1021/jm0608356
Jaghoori MM, Bleijlevens B, Olabarriaga SD (2016) 1001 Ways to run AutoDock Vina for virtual screening. J Comput Aided Mol Des 30:237–249. doi:10.1007/s10822-016-9900-9
Lang PT, Brozell SR, Mukherjee S et al (2009) DOCK 6: combining techniques to model RNA–small molecule complexes. RNA 15:1219–1230. doi:10.1261/rna.1563609
Meng EC, Shoichet BK, Kuntz ID (1992) Automated docking with grid-based energy evaluation. J Comput Chem 13:505–524. doi:10.1002/jcc.540130412
Zou X, Sun Y, Kuntz I (1999) Inclusion of solvation in ligand binding free energy calculations using the generalized-Born model. J Am Chem Soc 121:8033–8043. doi:10.1021/ja984102p
Liu HY, Kuntz ID, Zou XQ (2004) Pairwise GB/SA scoring function for structure-based drug design. J Phys Chem B 108:5453–5462. doi:10.1021/jp0312518
IBM Corporation (2010) IBM SPSS Statistics for Windows. IBM Corporation, Armonk
Munkres J (1957) Algorithms for the assignment and transportation problems. J Soc Ind Appl Math 5:32–38. doi:10.1137/0105003
Kuntz ID, Blaney JM, Oatley SJ et al (1982) A geometric approach to macromolecule–ligand interactions. J Mol Biol 161:269–288. doi:10.1016/0022-2836(82)90153-X
Accelrys Software Inc. (2015) Discovery Studio Modeling Environment, release 4.1. Accelrys Software Inc., San Diego
dos Santos IA (2012) Construção de uma base de dados para triagem virtual de potenciais inibidores da UDP-N-acetilglicosamina pirofosforilase do Moniliophthora perniciosa. Universidade Estadual de Feira de Santana
Sander T, Freyss J, von Korff M, Rufener C (2015) DataWarrior: an open-source program for chemistry aware data visualization and analysis. J Chem Inf Model 55:460–473. doi:10.1021/ci500588j
ChemAxon (2015) MarvinSketch. ChemAxon, Budapest
Stewart JJP (2013) Optimization of parameters for semiempirical methods VI: more modifications to the NDDO approximations and re-optimization of parameters. J Mol Model 19:1–32. doi:10.1007/s00894-012-1667-x
Stewart JJP (2012) MOPAC. Stewart Computational Chemistry, Colorado Springs
Balius TE, Mukherjee S, Rizzo RC (2011) Implementation and evaluation of a docking-rescoring method using molecular footprint comparisons. J Comput Chem 32:2273–89. doi:10.1002/jcc.21814
Brozell SR, Mukherjee S, Balius TE et al (2012) Evaluation of DOCK 6 as a pose generation and database enrichment tool. J Comput Aided Mol Des 26:749–773. doi:10.1007/s10822-012-9565-y
Gordon JC, Myers JB, Folta T et al (2005) H++: a server for estimating pK as and adding missing hydrogens to macromolecules. Nucleic Acids Res 33:W368–W371. doi:10.1093/nar/gki464
Myers J, Grothaus G, Narayanan S, Onufriev A (2006) A simple clustering algorithm can be accurate enough for use in calculations of pKs in macromolecules. Proteins Struct Funct Bioinf 63:928–938. doi:10.1002/prot.20922
Anandakrishnan R, Aguilar B, Onufriev AV (2012) H++ 3.0: Automating pK prediction and the preparation of biomolecular structures for atomistic molecular modeling and simulations. Nucleic Acids Res 40:537–541. doi:10.1093/nar/gks375
Morris GM, Huey R, Lindstrom W et al (2009) AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J Comput Chem 30:2785–2791. doi:10.1002/jcc.21256
Pettersen EF, Goddard TD, Huang CC et al (2004) UCSF Chimera? A visualization system for exploratory research and analysis. J Comput Chem 25:1605–1612. doi:10.1002/jcc.20084
Gasteiger J, Marsili M (1978) A new model for calculating atomic charges in molecules. Tetrahedron Lett 19:3181–3184. doi:10.1016/S0040-4039(01)94977-9
Gasteiger J, Marsili M (1980) Iterative partial equalization of orbital electronegativity—a rapid access to atomic charges. Tetrahedron 36:3219–3228. doi:10.1016/0040-4020(80)80168-2
Kitchen DB, Decornez H, Furr JR, Bajorath J (2004) Docking and scoring in virtual screening for drug discovery: methods and applications. Nat Rev Drug Discov 3:935–949. doi:10.1038/nrd1549
Mohammadi M, McMahon G, Sun L, et al. (1997) Structures of the tyrosine kinase domain of fibroblast growth factor receptor in complex with inhibitors. Science 276:955–960
Kinoshita T, Yoshida I, Nakae S et al (2008) Crystal structure of human mono-phosphorylated ERK1 at Tyr204. Biochem Biophys Res Commun 377:1123–1127. doi:10.1016/j.bbrc.2008.10.127
Williams NK, Bamert RS, Patel O et al (2009) Dissecting specificity in the Janus kinases: the structures of JAK-specific inhibitors complexed to the JAK1 and JAK2 protein tyrosine kinase domains. J Mol Biol 387:219–232. doi:10.1016/j.jmb.2009.01.041
Forli S, Huey R, Pique ME et al (2016) Computational protein–ligand docking and virtual drug screening with the AutoDock suite. Nat Protoc 11:905–919. doi:10.1038/nprot.2016.051
Nunes RR, dos Santos Costa M, dos Reis Santos B, et al (2016) Successful application of a virtual screening and molecular dynamics simulation against antimalarial molecular targets. Mem Inst Oswaldo Cruz 1–10. doi:10.1590/0074-02760160207
Habib E, Maia B, Campos VA, et al (2017) Octopus: a platform for the virtual high-throughput screening of a pool of compounds against a set of molecular targets. J Mol Model 23:26. doi: 10.1007/s00894-016-3184-9
Acknowledgements
The authors are grateful for the support provided by the Foundation for Research Support of Minas Gerais (FAPEMIG APQ-00557-14 and APQ-02860-16), the Higher Level Personnel Improvement Commission (CAPES), the National Research Council (CNPq UNIVERSAL 449984/2014-1), and the Graduate Program in Pharmaceutical Sciences from the Federal University of Sao Joao del Rei (PPGCF/UFSJ).
Author information
Authors and Affiliations
Corresponding author
Additional information
This paper belongs to Topical Collection Brazilian Symposium of Theoretical Chemistry (SBQT 2015)
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(DOCX 75 kb)
Rights and permissions
About this article
Cite this article
Carregal, A.P., Maciel, F.V., Carregal, J.B. et al. Docking-based virtual screening of Brazilian natural compounds using the OOMT as the pharmacological target database. J Mol Model 23, 111 (2017). https://doi.org/10.1007/s00894-017-3253-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00894-017-3253-8