Abstract
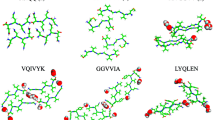
To date, there is little structural data available on the AGAAAAGA palindrome in the hydrophobic region (113–120) of prion proteins, although many experimental studies have shown that this region has amyloid fibril forming properties. This region belongs to the N-terminal unstructured region (1–123) of prions, the structure of which has proved hard to determine using NMR or X-ray crystallography. This paper reports the successful construction of three amyloid fibril models for this region. The models were formatted by standard simulated annealing using suitable templates from the Protein Data Bank, and were refined using several traditional optimization methods within AMBER. Because the NMR or X-ray structure of the hydrophobic region AGAAAAGA of prion proteins has not yet been determined, these models can be used as a reference for experimental studies on this region. The results presented here confirm standard simulated annealing as an effective tool in molecular modeling. The three constructed models for amyloid fibrils may be useful in furthering the goals of medicinal chemistry in this field.






Similar content being viewed by others
References
Prusiner SB (1982) Novel proteinaceous infectious particles cause scrapie. Science 216:136–144
Prusiner SB (1998) Prions. Proc Natl Acad Sci USA 95:13363–13383
Griffith JS (1967) Self-replication and scrapie. Nature 215:1043–1044
Brown DR (2000) Prion protein peptides: optimal toxicity and peptide blockade of toxicity. Mol Cell Neurosci 15:66–78
Brown DR (2001) Microglia and prion disease. Microsc Res Tech 54:71–80
Holscher C, Delius H, Burkle A (1998) Overexpression of nonconvertible PrPc delta114–121 in scrapie-infected mouse neuroblastoma cells leads to trans-dominant inhibition of wild-type PrPSc accumulation. J Virol 72:1153–1159
Jobling MF, Huang X, Stewart LR, Barnham KJ, Curtain C, Volitakis I, Perugini M, White AR, Cherny RA, Masters CL, Barrow CJ, Collins SJ, Bush AI, Cappai R (2001) Copper and zinc binding modulates the aggregation and neurotoxic properties of the prion peptide PrP 106–126. Biochemistry 40:8073–8084
Jobling MF, Stewart LR, White AR, McLean C, Friedhuber A, Maher F, Beyreuther K, Masters CL, Barrow CJ, Collins SJ, Cappai R (1999) The hydrophobic core sequence modulates the neurotoxic and secondary structure properties of the prion peptide 106–126. J Neurochem 73:1557–1565
Kuwata K, Matumoto T, Cheng H, Nagayama K, James TL, Roder H (2003) NMR-detected hydrogen exchange and molecular dynamics simulations provide structural insight into fibril formation of prion protein fragment 106–126. Proc Natl Acad Sci USA 100:14790–14795
Norstrom EM, Mastrianni JA (2005) The AGAAAAGA palindrome in PrP is required to generate a productive PrPSc-PrPC complex that leads to prion propagation. J Biol Chem 280:27236–27243
Wegner C, Romer A, Schmalzbauer R, Lorenz H, Windl O, Kretzschmar HA (2002) Mutant prion protein acquires resistance to protease in mouse neuroblastoma cells. J Gen Virol 83:1237–1245
Brown DR, Herms J, Kretzschmar HA (1994) Mouse cortical cells lacking cellular PrP survive in culture with a neurotoxic PrP fragment. Neuroreport 5:2057–2060
Zhang JP (2009) Studies on the structural stability of rabbit prion probed by molecular dynamics simulations. J Biomol Struct Dyn 27:159–162
Cosentino U, Pitea D, Moro G, Saracino GAA, Caria P, Var RM, Colombo L, Forloni G, Tagliavini F, Salmona M (2008) The anti-fibrillogenic activity of tetracyclineson PrP 106–126: a 3D-QSAR study. J Mol Model 14:987–994
Henriques ST, Pattenden LK, Aguilar MI, Castanho MA (2008) PrP (106–126) does not interact with membranes under physiological conditions. Biophys J 95:1877–1889
Okimoto N, Yamanaka K, Suenaga A, Hata M, Hoshimo T (2003) Molecular dynamics simulations of prion proteins—effect of Ala117 → Val mutation. Chem-Bio Informatics J 3:1–11
Villa A, Mark AE, Saracino GAA, Cosentino U, Pitea D, Moro G, Salmona M (2006) Conformational polymorphism of the PrP 106–126 peptide in different environments: a molecular dynamics study. J Phys Chem B 110:1423–1428
Zheng W, Wang L, Hong Y, Sha Y (2009) PrP 106–126 peptide disrupts lipid membranes influence of C-terminal amidation. Biochem Biophys Res Commun 379:298–303
Sawaya MR, Sambashivan S, Nelson R, Ivanova MI, Sievers SA, Apostol MI, Thompson MJ, Balbirnie M, Wiltzius JJ, McFarlane HT, Madsen A, Riekel C, Eisenberg D (2007) Atomic structures of amyloid cross-beta spines reveal varied steric zippers. Nature 447:453–457
Chen HF (2009) Aggregation mechanism investigation of the GIFQINS cross-β amyloid fibril. Comput Biol Chem 33:41–45
Lee SW, Mou Y, Lin SY, Chou FC, Tseng WH, Chen C, Lu CYD, Yu SSF, Chan JCC (2008) Steric zipper of the amyloid fibrils formed by residues 109 to 122 of the Syrian hamster prion protein. J Mol Biol 378:1142–1154
Simone AD, Pedone C, Vitagliano L (2008) Structure, dynamics, and stability of assemblies of the human prion fragment SNQNNF. Biochem Biophys Res Commun 366:800–806
Vitagliano L, Stanzione F, Simone AD, Esposito L (2009) Dynamics and stability of amyloid-like steric zipper assemblies with hydrophobic dry interfaces. Biopolymers 91:1161–1171
Yamaguchi K, Matsumoto T, Kuwata K (2008) Critical region for amyloid fibril formation of mouse prion protein: unusual amyloidogenic properties of the helix 2 peptide. Biochemistry 47:13242–13251
Zhang ZQ, Chen H, Bai HJ, Lai LH (2007) Molecular dynamics simulations on the oligomer formation process of the GNNQQNY peptide from yeast prion protein Sup35. Biophys J 93:1484–1492
Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE (2000) The protein data bank. Nucleic Acids Res 28:235–242
Case DA, Darden TA, Cheatham III TE, Simmerling CL, Wang J, Duke RE, Luo R, Crowley M, Walker RC, Zhang W, Merz KM, Wang B, Hayik S, Roitberg A, Seabra G, Kolossvry I, Wong KF, Paesani F, Vanicek J, Wu X, Brozell SR, Steinbrecher T, Gohlke H, Yang L, Tan C, Mongan J, Hornak V, Cui G, Mathews DH, Seetin MG, Sagui C, Babin V, Kollman PA (2008) AMBER 10. University of California, San Francisco. Amber tutorials: http://ambermd.org/tutorials/
Li X, Chen XD (2005) Global convergence of shortest-residual family of conjugate gradient methods without line search. Asia-Pacific J Oper Res 22:529–538
Sun J, Zhang JP (2001) Global convergence of conjugate gradient methods without line Search. Ann Oper Res 103:161–173
Zhu H, Chen XD (2008) Global convergence of a special case of the the Dai-Yuan family without line search. Asia-Pacific J Oper Res 25:411–420
Bagirov AM, Zhang JP (2003) Comparative analysis of the cutting angle and simulated annealing methods in global optimization. Optimization 52:363–378
Kirkpatrick S, Gelatt CD, Vecchi MP (1983) Optimization by simulated annealing. Science 220:671–680
Humphrey W, Dalke A, Schulten K (1996) VMD—visual molecular dynamics. J Mol Graph 14:33–38
Zhang ZQ, Chen H, Lai LH (2007) Identification of amyloid fibril-forming segments based on structure and residue-based statistical potential. Bioinformatics 23:2218–2225
Gossert AD, Bonjour S, Lysek DA, Fiorito F, Wuthrich K (2005) Prion protein NMR structures of elk and of mouse/elk hybrids. Proc Natl Acad Sci USA 102:646–650
Gorfe AA, Caflisch A (2007) Ser170 controls the conformational multiplicity of the loop 166–175 in prion proteins: implication for conversion and species barrier. FASEB J 21:3279–3287
Wiltzius JJW, Landau M, Nelson R, Sawaya MR, Apostol MI, Goldschmidt L, Soriaga AB, Cascio D, Rajashankar K, Eisenberg D (2009) Molecular mechanisms for protein-encoded inheritance. Nat Struct Mol Biol 16:973–978
Zhang JP (2004) Derivative-free hybrid methods in global optimization and their applications, PhD thesis, The University of Ballarat, Australia
Acknowledgments
The author thanks Dr. Zhuqing Zhang (Peking University, China) for his help in preparing Fig. 4. The author appreciates the Editor-in-Chief for his suggestions and the anonymous referees for their numerous insightful comments, which have greatly improved this paper. Last, but not least, thanks go to Dr. Judy-Anne Osborn of the Australian National University and staff of Springer (http://www.springer.com/) for their help in improving my English of this paper. This paper is dedicated to the memory of my PhD supervisor Professor Alex M. Rubinov; the hybrid global and local optimization search strategy [38] of this paper was learned from him and Professor Adil M. Bagirov, another PhD supervisor of mine.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Zhang, J. Optimal molecular structures of prion AGAAAAGA amyloid fibrils formatted by simulated annealing. J Mol Model 17, 173–179 (2011). https://doi.org/10.1007/s00894-010-0691-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00894-010-0691-y