Abstract
The location of 5S and 35S rDNA sequences in chromosomes of four Aconitum subsp. Aconitum species was analyzed after fluorescence in situ hybridization (FISH). Both in diploids (2n = 2x = 16; Aconitum variegatum, A. degenii) and tetraploids (2n = 4× = 32; A. firmum, A. plicatum), rDNA repeats were localized exclusively on the shorter arms of chromosomes, in subterminal or pericentromeric sites. All analyzed species showed similar basal genome size (Cx = 5.31–5.71 pg). The most striking features of tetraploid karyotypes were the conservation of diploid rDNA loci and emergence of many additional 5S rDNA clusters. Chromosomal distribution of excessive ribosomal sites suggests their role in the secondary diploidization of tetraploid karyotypes.
Similar content being viewed by others
Introduction
Almost all Aconitum species studied so far have an interesting bimodal karyotype (x = 8) with two large (metacentric and submetacentric) and six small (usually submetacentric) chromosomes (Yuan and Yang 2006, Hong et al. 2017). Most taxa are diploid (2n = 16) or tetraploid (2n = 32), but triploid, pentaploid, hexaploid, and octoploid plants have been also detected (Simon et al. 2001). Only diploid A. fletcherianum (possessing two large metacentric chromosome pairs and x = 6) has a different genome construction (Hong et al. 2016). It seems puzzling that during 30 million years of evolution of so widely distributed and species-rich genus (Jabbour and Renner 2012), its karyotype uniformity has been preserved.
Aconitum karyotypes can show marked differences in the amount of heterochromatin, especially at sites localized within bands associated with nuclear organizers. From three diploid taxa belonging to the subgenus Aconitum, A. degenii and A. lasiocarpum showed a heterochromatin-poor karyotype with relatively stable C-banding patterns, while A. variegatum generally showed a higher amount of heterochromatin (Joachimiak et al. 1999). Two high-mountain tetraploid species belonging to this subgenus, Western Carpathian A. firmum and Sudetic A. plicatum, also contained different amounts of heterochromatin (Mitka et al. 2007). This suggests that Aconitum karyotypes, although stable at the level of chromosome morphology, are differentiated at the intra-chromosomal level. The existence of such differences has much potential use in taxonomical and phytogeographical studies of Aconitum, especially because of the general lack of reproducible, species-specific molecular markers within this genus. It has been shown that many well-defined or geographically disjunct Aconitum taxa possess identical or nearly identical ITS (internal transcribed spacer) and cpDNA sequences (Kita et al. 1995; Kita and Ito 2000; Utelli et al. 2000; Luo et al. 2005; Mitka et al. 2016). Although some arbitrarily amplified sequences (RAPD, ISSR) seem to be useful for the estimation of molecular diversity of target Aconitum genomes (Fico et al. 2003; Zhang et al. 2005; Mitka et al. 2007), low reproducibility and the generation of dominant, non-locus-specific markers limit the broader applicability of these methods in phylogenetic studies.
Almost all the karyotypic data on Aconitum are restricted to chromosome number and morphology, DAPI/CMA fluorochrome staining, and C-banding patterns (Okada 1991; Joachimiak et al. 1999; Simon et al. 2001; Ilnicki 2005; Mitka et al. 2007). There is no information on the localization of marker sequences in chromosomes, molecular composition of heterochromatic bands, and nuclear genome size of this genus. All this information could be useful in the identification of chromosomes, evaluation of intra- and inter-specific diversity, and acquiring knowledge on the origin and evolution of polyploid species. In the present study, we analyzed karyotypes of two diploid (A. variegatum and A. degeni) and two tetraploid (A. firmum and A. plicatum) species of Aconitum belonging to the subgen. Aconitum by fluorescence in situ hybridization with 35S rDNA and 5S rDNA probes. Chromosomal localization of rDNA sequences, along with information on the C-banding patterns (Joachimiak et al. 1999; Ilnicki 2005; Mitka et al. 2007), was used for more precise characterization of karyotype in these species. Additionally, the genome size of analyzed species was established by flow cytometry.
Materials and methods
Plant material and chromosome preparation
The materials for the study were specimens of Aconitum variegatum L. and A. degenii Gáyer (2n = 2× = 16), A. firmum Rhb., and A. plicatum Köhler ex Rchb. (2n = 4× = 32) belonging to the subgenus Aconitum, collected from natural sites in Carpathians and Sudetes and then cultivated in the Botanical Garden of the Jagiellonian University in Kraków. All of the analyzed species are rare and protected in Poland. Three of them (A. degenii, A. firmum, and A. plicatum) are listed in the Polish Red Data Book of Plants (2001) and classified as vulnerable species. A. variegatum is a Central European endemic representing the mountain element in the lowlands, and A. degenii is an endemic of eastern and southern Carpathians. From the two high-mountain tetraploid taxa, A. firmum is a western Carpathian endemic, and A. plicatum is a central European endemic known in Poland from the Sudetes. Three plants of Polish origin from each species were analyzed by FISH. Cytometric DNA estimations were performed not only on these plants but also on material collected in other Carpathian sites (Table 1).
Plants were taken to the laboratory and kept in pots, and then tips of roots were cut from them for cytological analysis. The material was pretreated with saturated aqueous solution of α-bromonaphtalene overnight at 4 °C and fixed in absolute ethanol/glacial acetic acid. After fixation, root tips were macerated with a 1% mixture of pectinase and cellulase for 10 min at 37 °C. Preparations were made by squashing in a drop of 45% acetic acid. Cover slips were then flicked off after freezing in dry ice, air-dried, and kept at − 24 °C until used.
DNA probe labeling and FISH
The procedure followed the protocol described in detail by Wolny and Hasterok (2009) with minor modifications. In brief, the following DNA probes were used: (i) a 2.3 kb ClaI subclone representing the fragment of a 25S rDNA genic region of Arabidopsis thaliana (Unfried and Gruendler 1990) was labeled by nick translation with digoxigenin-11-dUTP (Roche, Indianapolis, IN, USA) and used to detect the 35S rDNA loci containing the genes encoding for 18S-5.8S-25S rRNA. (ii) The pTa794 clone containing a 410 bp fragment of 5S rDNA unit isolated from wheat (Gerlach and Dyer 1980) was labeled by PCR with tetramethyl-rhodamime-5-dUTP (Roche) and used to visualize 5S rDNA loci. Primer sequences and conditions for the reactions were as described by Hasterok et al. (2002).
General conditions of the FISH procedure were as follows: slides were incubated with DNase-free RNase (100 μg/ml) in 2× SSC for 1 h at 37 °C, then washed in three changes of 2× saline sodium citrate (SSC) buffer for 15 min, post-fixed in 1% formaldehyde in PBS buffer followed by washes in 2× SSC for 15 min, dehydrated in an ethanol series (70, 90, and 100%) and air-dried. The DNA probes were mixed to a final concentration of 2.5–3.5 ng/μl of hybridization mixture along with 50% deionized formamide, 20% dextran sulfate, 2× SSC, 0.5% SDS, and salmon sperm blocking DNA in 500–100× excess of labeled probes. The hybridization mixture was pre-denatured (75 °C for 10 min), applied to the chromosome preparations, and denatured together at 75 °C for 5 min in an Omnislide in situ hybridization system (Hybaid, Basingstoke, UK) and then incubated overnight at 37 °C in a humid chamber to allow renaturation. After hybridization, slides were washed for 10 min in 15% deionized formamide in 0.1× SSC at 42 °C, which is equivalent to 82% stringency, followed by several washes in 2× SSC. Digoxigenated probe was immunodetected according to standard protocols by antidigoxigenin antibodies conjugated with fluorescein isothiocyanate (FITC; Roche). After final dehydration, preparations were mounted and counter-stained in Vectashield (Vector Laboratories, Burlingame, CA, USA) antifade buffer containing 2.5 μg/ml 4′-6-diamidino-2-phenylindole (DAPI; Serva). Images were taken using a Leica DMRB epifluorescent microscope equipped with a CoolSnap (Photometrics) monochromatic CCD camera and then processed uniformly using Picture Publisher (Micrographx) software. For visualization of very tiny FISH signals, we used the option “find edges” of the ImageJ program (http://rsb.info.nih.gov/ij/).
The karyograms with marked FISH-detected rDNA segments were constructed on the basis of the generalized karyotype (Table 2) of four Aconitum species. It was obtained from the measurements of 40 DAPI-stained metaphases (10 per each species). For the heteromorphic pairs/groups of chromosomes, additive chromatin segments (0.2–0.3 μm long) were taken into account.
Nuclear DNA content measurements
For the flow cytometric estimation of nuclear DNA content, Pisum sativum cv. Set (2C = 9.11 pg; Sliwinska et al. 2005) was used as an internal standard. Young leaves derived from adult plants were used as material for the study. Plant tissues of Aconitum and P. sativum were chopped simultaneously with a sharp razor blade in a plastic Petri dish with 1 ml nucleus-isolation buffer LB01 (Doležel et al. 1989), supplemented with propidium iodide (50 μg/ml) and ribonuclease A (50 μg/ml). After chopping, the suspension was passed through a 50-μm mesh nylon filter. For each sample, 6000–10,000 nuclei were analyzed directly after preparation using a Partec CCA flow cytometer (Münster, Germany). Analyses were replicated three times for each specimen. Histograms were analyzed using a DPAC v.2.2 computer program. Absolute nuclear DNA content was calculated using the linear relationship between the ratios of the 2C peak positions Aconitum/Pisum, on the histogram of fluorescence intensities (Galbraith et al. 1998).
Results
Diploids
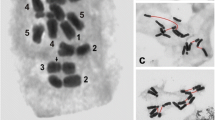
The two diploid species showed similar distribution of rDNA sites within the karyotype. All detected 35S rDNA sequences were localized terminally and 5S rDNA sequences pericentromerically. There were eight 35S rDNA signals within the chromosome complement of A. variegatum, localized on the shorter arms of four-chromosome pairs (1, 3, 4, and 5) (Figs. 1a, b and 2a). In A. degenii, only six 35S rDNA sites were detected. Four of them were localized on chromosomes 3 and 5, one on chromosome 1 and one on chromosome 7 (Figs. 1c, d and 2b). 35S rDNA clusters on chromosomes 1, 3, and 5 seem to be the major nucleolar-organizing regions (NORs) in the analyzed plants. A specific feature of the A. variegatum karyotype was the occurrence of two distinct 35S rDNA signals in the non-satellited chromosome pair 4 (Fig. 2a).
Fluorescence in situ hybridization of 35S rDNA (green) and 5S rDNA (red) probes to the somatic metaphase chromosomes of a, b A. variegatum and c, d A. degenii. Images b and d were processed with the use of the option “find edges” of the ImageJ program. The chromosomes were counter-stained with DAPI (blue). Arrows indicate small 5S rDNA site in chromosome 5, detected after image processing. Bar 5 μm
Karyotype structure of A. variegatum (a) and A. degenii (b). Green—35S rDNA, red—5S rDNA, black—fixed heterochromatin segments, gray—polymorphic heterochromatin segments (heterochromatin classification according to Joachimiak et al. 1997). The position of satellites and heterochromatin was taken from Joachimiak et al. (1999), Ilnicki (2005), and Mitka et al. (2007). Bar 5 μm
Both A. variegatum and A. degenii showed two large 5S rDNA sites localized within the pericentromeric heterochromatin of the shorter arms of the longest chromosomes (pair 1) (Figs. 1 and 2). Additionally, chromosome 5 may harbor a very tiny 5S rDNA signal near centromere, practically undetectable without special image processing. Only one chromosome 5 with such signal was detected in both species, indicating that 5S rDNA repeats were absent or at a copy number below detection level on the homolog.
The nuclear 2C DNA amount was estimated at 11.43 pg in A. variegatum and 11.23 pg in A. degenii, so the basal genome size is nearly identical in both specie (5.71 and 5.61 pg, respectively) (Table 3).
Tetraploids
The two tetraploid species analyzed here showed a high number of 35S and 5S rDNA loci (Figs. 3 and 4). However, some rDNA loci were small and thus not detectable in all preparations. The distribution of 5S rDNA signals was similar to that observed in diploids: almost all detected sequences were located pericentromerically. 35S rDNA clusters were located not only terminally but also pericentromerically. In all instances, pericentromeric clusters of 35S rDNA colocalized with 5S rDNA (Figs. 5 and 6).
The number of detected 35S rDNA signals was up to 20 in A. firmum and up to 14 in A. plicatum. The most characteristic feature of Carpathian A. firmum was the heterozygosity of the first and third four-chromosome group in respect of the 35S rDNA distribution (Fig. 5). The karyotype of this species showed major 35S rDNA clusters on the ends of two chromosomes of type 1 and in the pericentromeres of two satellited chromosomes of type 3. The same chromosome sites in Sudetic A. plicatum were invariably lacking 35S rDNA signals (Fig. 6).
When compared to the two diploid species, the number of 35S rDNA loci in A. plicatum (14) did not show clear deviation from additivity. A. firmum showed, however, more 35S rDNA signals (20) than expected from the karyotypes of the two analyzed Carpathian diploids (6–8).
The most striking feature of the tetraploid karyotypes is the presence of a huge number of 5S rDNA clusters, located in chromosomes belonging to all morphological groups. Both analyzed tetraploids showed a similar pericentromeric distribution of these repeats. The number of detected 5S rDNA signals was up to 23 in A. firmum and up to 20 in A. plicatum. Many of these signals were small and hardly detectable. The most prominent were located in the pericentromeric region of the shorter arms of chromosomes 1 (4 signals), 4 (2 signals), and 8 (2 signals).
The nuclear DNA content estimated for A. firmum was 2C = 21.62 pg and 2C = 21.24 pg for A. degenii, so the basal genome size is nearly identical in both species (Cx = 5.40 pg and Cx = 5.31 pg, respectively) (Table 3).
Discussion
In this paper, we presented for the first time a thorough FISH analysis of the rDNA sites in Aconitum. Together with the previously performed chromosome studies, it provides interesting data on the karyotype structure and evolution in this species-rich and widely spread genus.
The majority of terminal 35S rDNA sites detected in analyzed species by FISH were frequently observed as small satellites in conventionally stained preparations (Joachimiak et al. 1999; Ilnicki 2005). A similar localization was reported for chromomycin A3-positive and DAPI-negative (CMA+/DAPI−) chromatin segments in the Japanese diploid species A. sanyoense (Okada 1991). In the majority of plant species, CMA+/DAPI− staining is characteristic for NOR-associated chromatin (Guerra 2000), so it can be deduced that the distribution of major NOR loci was the same in all three diploid species. According to Roa and Guerra (2012), the stability of rDNA sites is common phenomenon within taxa characterized by conserved chromosome morphology.
The terminal 35S rDNA sites were also observed on the non-satellited chromosome pair 4 in A. variegatum. The presence of 35S rDNA repeats has frequently been reported in regions where secondary constrictions have not been located (Guerra 2000, Roa and Guerra 2012). Most likely, they represent inactive, epigenetically silenced rDNA sites. Such silencing of rDNA clusters located out of NOR chromosomes was demonstrated in Rumex acetosa where 35S rDNA on the non-satellited chromosome pair was highly methylated (Lengerova and Vyskot 2001).
All terminally located 35S rDNA sites detected here were previously described as C-band positive (Joachimiak et al. 1999; Ilnicki 2005). They constituted (together with the major 5S rDNA cluster localized near centromere of chromosome 1) the major heterochromatic loci in C-banded Aconitum genomes. Although different plants belonging to the analyzed taxa showed more C-band-positive segments, only some of these segments were conserved (i.e., observed in all or nearly all chromosomes of a given type). The most stable were those located at the centromeres and NORs. Interestingly, polymorphic heterochromatin in Aconitum does not generally contain rDNA, and all minor rDNA sites in diploids as well as minor and extra rDNA sites in tetraploids were not heterochromatinized.
A. degenii chromosome pair 1, detected here as heterozygous in respect of the presence of 35S rDNA on the end of the shorter arm, was also heterozygous in respect of the amount of heterochromatin at this position (Fig. 2b). Heterochromatin difference between homologs was stable and observed in many plants collected from very distant sites in Poland, the Ukraine, and Romania (Joachimiak et al. 1999; Ilnicki 2005). The same phenomenon was observed in A. lasiocarpum, a close relative of A. degenii (Joachimiak et al. 1999). The reason why plants with homomorphic pair 1 were not observed in analyzed material remains unknown. The absence of 35S rDNA in one chromosome of type 1 and the presence of this sequence in chromosome 7 in A. degenii suggested a 1:7 translocation of nucleolar-organizing region (Fig. 2b). The analysis of meiosis in plants belonging to the sect. Lycoctonum performed many years ago by Afify (1933) showed that chromosome translocations are not uncommon in Aconitum taxa. The structural heterozygosity of the first chromosome group was also observed in tetraploid A. firmum, both in FISH and C-banding studies.
A high number of rDNA sites observed in the analyzed tetraploids is a very interesting phenomenon because the general trend in chromosome evolution seems rather to be partial reduction of rDNA loci in polyploid species (Maluszynska and Heslop-Harrison 1993; Volkov et al. 1999; Krishnan et al. 2001; Mishima et al. 2002; Weiss-Schneeweiss et al. 2007; Roa and Guerra 2012). The important steps in the lowering of such loci were the inactivation of one of the parental rDNA and a gradual decrease in the number of rDNA repeats. Such changes, resulting in asymmetry or loss of rDNA clusters, were suggested in Brassica (Hasterok et al. 2006), Trifolium (Ansari et al. 2008), and Nicotiana (Kovarik et al. 2008). Variation in the number and position of rDNA repeats could result from the changes of chromosomes such as ectopic (interlocus) recombination, inversions and translocations, or transposition (Gernand et al. 2007). It has been suggested that the first two events are influenced by chromosomal location: terminal localization allows frequent rearrangement to occur without disrupting other gene linkages (Hanson et al. 1996). This is supported by observations in Gossypium and Nicotiana where sub-telomeric 35S rDNA loci show interlocus homogenization, while interstitial 5S rDNA loci do not (Cronn et al. 1996; Fulnecek et al. 2002). The lack of detectable changes in chromosome morphology and uniform pericentromeric location of 5S rDNA in tetraploid Aconitum plants suggest rather the involvement of transposition and further amplification of transposed rDNA sequences. The ability of rDNA sequences to transpose was reported by Schubert and Wobus (1985), Adams et al. (2000), Raskina et al. (2004), Datson and Murray (2006). It was supposed that mobility of repeated sequences may result from activation of dormant transposons by different stresses, including polyploidy and in vitro culture (Capy et al. 2000; Liu and Wendel, 2002; Gernand et al. 2007; and references cited therein). They can also be incorporated into transposable elements, as it was revealed specifically for 5S rDNA by Kalendar et al. (2008) or directly amplified as a satellite (Martins et al. 2006). The role of massive amplification of rDNA loci, observed only in some plant species, remains unknown. Most probably, it enhances the differences between similarly shaped chromosome pairs, as in Tulipa (Mizuochi et al. 2007), or reinforces the chromosome-arm homology, as in permanent translocation heterozygote Rhoeo spathacea (Golczyk et al. 2005). The rDNA distribution in tetraploid Aconitum species suggests the role of additional ribosomal sites in differentiation of chromosome pairs in these plants: in many instances, two chromosomes within a four-chromosome group showed the rDNA segment while two did not (Figs. 5 and 6).
The massive rDNA amplification in A. firmum and A. plicatum should result in a detectable increase of the genome size in both species, but Cx DNA values calculated for the analyzed species (5.71 and 5.61 pg in diploids; 5.40 and 5.31 pg in tetraploids) suggest instead a small reduction of the basal genome size in the tetraploids (Table 3). Genome downsizing is a well-known phenomenon detected in many polyploid plants (for review see Leitch and Bennett 2004), although in some plant genera, no discernible pattern in this direction has been observed (Grabowska-Joachimiak et al. 2006; Leitch et al. 2008; Klos et al. 2009; Anamthawat-Jónsson et al. 2010; Morozowska et al. 2015; Podwyszyńska et al. 2016). Most probably, global changes in genome size result from both the elimination and accumulation of different genomic sequences. Evidently, in tetraploid Aconitum species, the accumulation of rDNA repeats in new chromosomal sites was counterbalanced by the elimination of some other sequences from the genome. This may suggest the existence of mechanism that stabilizes the basal genome size in Aconitum. It is worth mentioning that the 1Cx value (5.44 pg) in Aconitum vulparia belonging to the subgen. Lycoctonum (the only Aconitum species analyzed so far in this respect: Siljak-Yakovlev et al. 2010) is located within the range of values given here (5.31–5.71 pg).
The gain or loss of different DNA sequences and reorganization of chromosomal loci in polyploids are parts of process called diploidization (Wolfe 2001). From a cytological point of view, it leads to the formation of well-differentiated chromosome pairs. This allows, among other advantages, to avoid pairing problems at meiosis and increase fertility. Although the diploidization subsequent to polyploidization plays an irrefutable role in releasing of evolutionary potential of newly formed polyploids, it still remains an under-studied topic in plants (Dodsworth et al. 2016).
References
Adams SP, Leitch IJ, Bennett MD, Chase MW, Leitch AR (2000) Ribosomal DNA evolution and phylogeny in Aloe (Asphodelaceae). Am J Bot 87:1578–1583
Afify A (1933) Chromosome form and behaviour in diploid and triploid Aconitum. J Genet 27:293–318
Anamthawat-Jónsson K, Thórsson ÆT, Temsch EM, Greilhuber J (2010) Icelandic birch polyploids—the case of perfect fit in genome size. J Bot Volume 2010, Article ID 347254, 9 pages. https://doi.org/10.1155/2010/347254
Ansari HA, Ellison NW, Williams WM (2008) Molecular and cytogenetic evidence for an allotetraploid origin of Trifolium dubium (Leguminosae). Chromosoma 117:159–167. https://doi.org/10.1007/s00412-007-0134-4
Capy P, Gasperi G, Biemont C, Bazin C (2000) Stress and transposable elements: co-evolution or useful parasites? Heredity 85:101–106. https://doi.org/10.1046/j.1365-2540.2000.00751.x
Cronn RC, Zhao X, Paterson AH, Wendel JF (1996) Polymorphism and concerted evolution in a tandemly repeated gene family: 5S ribosomal DNA in diploid and allopolyploid cottons. J Mol Evol 42:685–705. https://doi.org/10.1007/BF02338802
Datson PM, Murray BG (2006) Ribosomal DNA locus evolution in Nemesia: transposition rather than structural rearrangement as the key mechanism? Chromosom Res 14:845–857. https://doi.org/10.1007/s10577-006-1092-z
Dodsworth S, Chase MW, Leith AR (2016) Is post-polyploidization dipoidization the key to the evolutionary success of angiosperms ? Bot J Linn Soc 180(1):1–5. https://doi.org/10.1111/boj.12357
Doležel J, Binarová P, Lucretti S (1989) Analysis of nuclear DNA content in plant cells by flow cytometry. Biol Plantarum 31:113–120. https://doi.org/10.1007/BF02907241
Fico G, Spada A, Braca A, Agradi E, Morelli I, Tomé F (2003) RAPD analysis and flavonoid composition of Aconitum as an aid for taxonomic discrimination. Biochem Syst Ecol 31:293–301
Fulnecek J, Lim KY, Leitch AR, Kovarik A, Matyasek R (2002) Evolution and structure of 5S rDNA loci in allotetraploid Nicotiana tabacum and its putative parental species. Heredity 88:19–25. https://doi.org/10.1038/sj.hdy.6800001
Galbraith DW, Lambert GM, Macas J, Doležel J (1998) Analysis of nuclear DNA content and ploidy in higher plants. In: Robinson JP, Darzynkiewicz Z, Dean PN, Dressler LG, Orfao A, Rabinovitch PS, Stewart CC, Tanke HJ, Wheeless LL (eds) Current protocols in cytometry. John Wiley & Sons Inc., New York, pp 7.6.1–7.6.22. https://doi.org/10.1002/0471142956.cy0706s02
Gerlach WL, Dyer TA (1980) Sequence organization of the repeating units in the nucleus of wheat which contain 5S rRNA genes. Nucleic Acids Res 8:4851–4865. https://doi.org/10.1093/nar/8.21.4851
Gernand D, Golczyk H, Rutten T, Ilnicki T, Houben A, Joachimiak AJ (2007) Tissue culture triggers chromosome alterations, amplification, and transposition of repeat sequences in Allium fistulosum. Genome 50:435–442. https://doi.org/10.1139/g07-023
Golczyk H, Hasterok R, Joachimiak AJ (2005) FISH-aimed karyotyping and characterization of Renner complexes in permanent heterozygote Rhoeo spathacea. Genome 48:145–153. https://doi.org/10.1139/g04-093
Grabowska-Joachimiak A, Sliwinska E, Piguła M, Skomra U, Joachimiak AJ (2006) Genome size in Humulus lupulus L. and H. japonicus Siebold & Zucc. (Cannabaceae). Acta Soc Bot Pol 75:207–214
Guerra M (2000) Patterns of heterochromatin distribution in plant chromosomes. Genet Mol Biol 23:1029–1041. https://doi.org/10.1590/S1415-47572000000400049
Hanson RE, Islam-Faridi MN, Percival EA, Crane CF, Ji Y, McKnight TD, Stelly DM, Price HJ (1996) Distribution of 5S and 18S-25S rDNA loci in tetraploid cotton (Gossypium hirsutum L.) and its putative diploid ancestors. Chromosoma 105:55–61. https://doi.org/10.1007/BF02510039
Hasterok R, Langdon T, Taylor S, Jenkins G (2002) Combinatorial labelling of DNA probes enables multicolour fluorescence in situ hybridisation in plants. Folia Histochem Cytobiol 40:319–323
Hasterok R, Wolny E, Hosiawa M, Kowalczyk M, Kulak-Ksiazczyk S, Ksiazczyk T, Heneen W, Maluszynska J (2006) Comparative analysis of rDNA distribution in chromosomes of various species of Brassicaceae. Ann Bot 97:205–216. https://doi.org/10.1093/aob/mcj031
Hong Y, Gao Q, Luo Y, Luo JP, Zhang Y, Yuan Q, Yang QE (2016) Karyology of Aconitum subgenus Lycoctonum (Ranunculaceae) from China, with a report of the new base chromosome number x = 6 in the genus Aconitum. Nord J Bot 34:441–454
Hong Y, Luo Y, Gao Q, Ren C, Yuan Q, Yang Q-E (2017) Phylogeny and reclassification of Aconitum subgenus Lycoctonum (Ranunculaceae). PLoS One 12(1):e0171038. https://doi.org/10.1371/journal.pone.0171038
Ilnicki T (2005) Chromosome variation of Aconitum variegatum L. in the Carpathians. Ph.D. thesis, Jagiellonian University, Institute of Botany, pp. 1–87 (in Polish)
Jabbour F, Renner SS (2012) A phylogeny of Delphinieae (Ranunculaceae) shows that Aconitum is nested with Delphinium and that late Miocene transitions to long life cycles in the Himalayas and Southwest China coincide with bursts in diversification. Mol Phylogenet Evol 62:928–942. https://doi.org/10.1016/j.ympev.2011.12.005
Joachimiak A, Kula A, Grabowska-Joachimiak A (1997) On heterochromatin in karyosystematic studies. Acta Biol Cracov Ser Bot 39:69–77
Joachimiak A, Ilnicki T, Mitka J (1999) Karyological studies on Aconitum lasiocarpum (Rchb.) Gayer (Ranunculaceae). Acta Biol Cracov Ser Bot 41:205–211
Kalendar R, Tanskanen J, Chang W, Antonius K, Sela H, Peleg O, Schulman AH (2008) Cassandra retrotransposons carry independently transcribed 5S RNA. Proc Natl Acad Sci USA 105:5833–5838. https://doi.org/10.1073/pnas.0709698105
Kita Y, Ito M (2000) Nuclear ribosomal ITS sequences and phylogeny in east Asian Aconitum subgenus Aconitum (Ranunculaceae), with special reference to extensive polymorphism in individual plants. Plant Syst Evol 225:1–13. https://doi.org/10.1007/BF00985455
Kita Y, Ueda K, Kadota Y (1995) Molecular phylogeny and evolution of the Asian Aconitum sugen. Aconitum (Ranunculaceae). J Plant Res 108:429–442. https://doi.org/10.1007/BF02344231
Klos J, Sliwinska E, Kula A, Golczyk H, Grabowska-Joachimiak A, Ilnicki T, Szostek K, Stewart A, Joachimiak AJ (2009) Karyotype and nuclear DNA content of hexa-, octo-, and duodecaploid lines of Bromus subgen. Ceratochloa. Genet Mol Biol 32:528–537. https://doi.org/10.1590/S1415-47572009005000046
Kovarik A, Dadejova M, Lim YK, Chase MW, Clarkson JJ, Knapp S, Leitch AR (2008) Evolution of rDNA in Nicotiana allopolyploids: a potential link between rDNA homogenization and epigenetics. Ann Bot 101:815–823. https://doi.org/10.1093/aob/mcn019
Krishnan P, Sapra VT, Soliman KM, Zipf A (2001) FISH mapping of the 5S and 18S-28S rDNA loci in different species of Glycine. J Hered 92:295–300. https://doi.org/10.1093/jhered/92.3.295
Leitch IJ, Bennett MD (2004) Genome downsizing in polyploid plants. Biol J Linn Soc 82:651–663. https://doi.org/10.1111/j.1095-8312.2004.00349.x
Leitch IJ, Hanson L, Lim KY, Kovarik A, Chase MW, Clarkson JJ, Leitch AR (2008) The ups and downs of genome size evolution in polyploid species of Nicotiana (Solanaceae). Ann Bot 101:805–814. https://doi.org/10.1093/aob/mcm326
Lengerova M, Vyskot B (2001) Sex chromatin and nucleolar analyses in Rumex acetosa L. Protoplasma 217:147–153. https://doi.org/10.1007/BF01283395
Liu B, Wendel JF (2002) Non-Mendelian phenomena in allopolyploid genome evolution. Curr Genomics 3:489–505. https://doi.org/10.2174/1389202023350255
Luo Z, Zhang FM, Yang Q-E (2005) Phylogeny of Aconitum subgenus Aconitum (Ranunculaceae) inferred from ITS sequences. Plant Syst Evol 252:11–25. https://doi.org/10.1007/s00606-004-0257-5
Maluszynska J, Heslop-Harrison P (1993) Physical mapping of rDNA loci in Brassica species. Genome 36:774–781. https://doi.org/10.1139/g93-102
Martins C, Ferreira IA, Oliveira C, Foresti F, Galetti PM Jr (2006) A tandemly repetitive centromeric DNA sequence of the fish Hoplias malabaricus (Characiformes: Erythrinidae) is derived from 5S rDNA. Genetica 127:133–141. https://doi.org/10.1007/s10709-005-2674-y
Mishima M, Ohmido N, Fukui K, Yahara T (2002) Trends in site-number change of rDNA loci during polyploid evolution in Sanguisorba (Rosaceae). Chromosoma 110:550–558. https://doi.org/10.1007/s00412-001-0175-z
Mitka J (2003) The genus Aconitum L. (Ranunculaceae) in Poland and adjacent countries. A phenetic study. The Institute of Botany of the Jagiellonian University, Kraków
Mitka J, Sutkowska A, Ilnicki T, Joachimiak AJ (2007) Reticulate evolution of high-alpine Aconitum (Ranunculaceae) in the western Sudetes and western Carpathians Central Europe. Acta Biol Cracov Ser Bot 49(2):15–26
Mitka J, Boroń P, Novikoff A, Wróblewska A, Binkiewicz B (2016) Two major groups of chloroplast DNA haplotypes in diploid and tetraploid Aconitum subgen. Aconitum (Ranunculaceae) in the Carpathians. Modern Phytomorphology 9(Suppl):5–15
Mizuochi H, Marasek A, Okazaki K (2007) Molecular cloning of Tulipa fosteriana rDNA and subsequent FISH analysis yields cytogenetic organization of 5S rDNA and 45S rDNA in T. gesneriana and T. fosteriana. Euphytica 155:235–248. https://doi.org/10.1007/s10681-006-9325-y
Morozowska M, Czarna A, Jędrzejczyk I, Bocianowski J (2015) Genome size, leaf, fruit and seed traits—taxonomic tools for species identification in the genus Nasturtium R. Br Acta Biol Cracov Bot 57(1):114–124. https://doi.org/10.1515/abcsb-2015-0011
Okada H (1991) Correspondence of Giemsa C-band with DAPI/CMA fluorochrome staining pattern in Aconitum sanyoense (Ranunculaceae). Cytologia 56:135–141
Podwyszyńska M, Kruczyńska D, Machlańska A, Dyki B, Sowik I (2016) Nuclear DNA content and ploidy level of apple cultivars including Polish ones in relation to some morphological traits. Acta Biol Cracov Ser Bot 58(1):81–93. https://doi.org/10.1515/abcsb-2016-0008
Polish Red Data Book of Plants (2001) Pteridophytes and flowering plants. In: Kaźmierczakowa R, Zarzycki K (eds) Polish Academy of Sciences. W. Szafer Institute of Botany, Institute of Nature Conservation, Kraków
Raskina O, Belyayev A, Nevo E (2004) Activity of the En/Spm-like transposons in meiosis as a base for chromosome repattering in a small, isolated, peripheral population of Aegilops speltoides Taush. Chromosom Res 12:153–161. https://doi.org/10.1023/B:CHRO.0000013168.61359.43
Roa F, Guerra M (2012) Distribution of 45S rDNA sites in chromosomes of plants: structural and evolutionary implications. BMC Evol Biol 12:225. https://doi.org/10.1186/1471-2148-12-225
Schubert I, Wobus U (1985) In-situ hybridization confirms jumping nucleolus organizing regions in Allium. Chromosoma 92:143–148. https://doi.org/10.1007/BF00328466
Siljak-Yakovlev S, Pustahija F, Solic EM, Bogunic F, Muratovic E, Basic N, Catrice O, Brown SC (2010) Towards a genome size and chromosome number database of Balkan flora: C-values in 343 taxa with novel values for 242. Adv Sci Lett 3:190–213. https://doi.org/10.1166/asl.2010.1115
Simon J, Bosch M, Molero J, Blanche C (2001) A conspect of chromosome numbers in tribe Delphinieae (Ranunculaceae). Available at http://www.ub.es/botanica/greb/cromodel.pdf
Sliwinska E, Zielinska E, Jedrzejczyk I (2005) Are seeds suitable for flow cytometric estimation of plant genome size? Cytometry Part A 64A:72–79. https://doi.org/10.1002/cyto.a.20122
Unfried I, Gruendler P (1990) Nucleotide sequence of the 5.8S and 25S rRNA genes and of the internal transcribed spacers from Arabidopsis thaliana. Nucleic Acids Res 18:4011. https://doi.org/10.1093/nar/18.13.4011
Utelli AB, Roy BA, Baltisbereger M (2000) Molecular and morphological analyses of European Aconitum species (Ranunculaceae). Plant Syst Evol 224:195–212. https://doi.org/10.1007/BF00986343
Volkov RA, Borisjuk NV, Panchuk II, Schweizer D, Hemleben V (1999) Elimination and rearrangement of parental rDNA in the allotetraploid Nicotiana tabacum. Mol Biol Evol 16:311–320. https://doi.org/10.1093/oxfordjournals.molbev.a026112
Weiss-Schneeweiss H, Schneeweiss GM, Stuessy TF, Mabuchi T, Park J-M, Jang C-G, Sun B-Y (2007) Chromosomal stasis in diploids contrasts with genome restructuring in auto- and allopolyploid taxa of Hepatica (Ranunculaceae). New Phytol 174:669–682. https://doi.org/10.1111/j.1469-8137.2007.02019.x
Wolfe KH (2001) Yesterday’s polyploids and the mystery of diploidization. Nat Rev Genet 2:333–341. https://doi.org/10.1038/35072009
Wolny E, Hasterok R (2009) Comparative cytogenetic analysis of the genomes of the model grass Brachypodium distachyon and its close relatives. Ann Bot 104:873–881. https://doi.org/10.1093/aob/mcp179
Yuan Q, Yang Q-E (2006) Polyploidy in Aconitum subgen. Lycoctonum (Ranunculaceae). Bot J Linn Soc 150:343–353. https://doi.org/10.1111/j.1095-8339.2006.00468.x
Zhang F-M, Chen W-L, Yang Q-E, Ge S (2005) Genetic differentiation and relationship of populations in the Aconitum delavayi complex (Ranunculaceae) and their taxonomic implications. Plant Syst Evol 254:39–48. https://doi.org/10.1007/s00606-005-0298-4
Acknowledgements
The authors thank Dr. Tim Langdon (IBERS, Aberystwyth University, UK) for his comments on the manuscript, Dr. Tomasz Ilnicki for the squashes on which FISH reactions were made, and Dr. Józef Mitka (Botanical Garden, Jagiellonian University) for supplying the plant material.
Funding
The work was supported by the State Committee for Scientific Research (Grant No. 2 P04C 072 29).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Handling Editor: Heiti Paves
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Joachimiak, A.J., Hasterok, R., Sliwinska, E. et al. FISH-aimed karyotype analysis in Aconitum subgen. Aconitum reveals excessive rDNA sites in tetraploid taxa. Protoplasma 255, 1363–1372 (2018). https://doi.org/10.1007/s00709-018-1238-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00709-018-1238-9