Abstract
A novel dsRNA virus named “Thelonectria quadrivirus 1” (TQV1) was found in a member of the genus Thelonectria (Ascomycota), isolated from a root associated with stem collar necrosis of Fraxinus excelsior L. The complete genome of TQV1 is composed of four segments, each containing a single ORF on the positive sense RNA. The sequence of the 5´ (5´-(C/T)ACGAAAAA-3´) and 3´termini (5´AT(T/G)AGCAATG(T/C)GC(G/A)CG-3’) of dsRNA 1 (4876 bp), dsRNA 2 (4312 bp), dsRNA 3 (4158 bp), and dsRNA 4 (3933 bp) are conserved. Based on its genome organization and phylogenetic position, TQV1 is suggested to be a new member of the family Quadriviridae. This is the first report of a mycovirus infecting a member of the genus Thelonectria.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
The most widespread mycoviruses are viruses with a dsRNA genome. According to Kotta-Loizou et al. [1], dsRNA mycoviruses are arranged in seven families (Totiviridae, Partitiviridae, Megabirnaviridae, Chrysoviridae, Quadriviridae, Endornaviridae and Reoviridae) and one genus (Botybirnavirus). At present, the family Quadriviridae consists of the single genus Quadrivirus. Within this genus, only one member, Rosellinia necatrix quadrivirus 1-W1075, has been confirmed so far [2, 3], but several other viruses have been proposed to belong to the family Quadriviridae.
The quadrivirus genome consists of four dsRNA segments, which are packed in non-enveloped spherical particles, 45 nm in diameter. The particles are composed of the structural proteins P2 and P4 and enclose the RdRP, which is encoded on segment 3. Segment 1 encodes a hypothetical protein with unknown function. The dsRNA segments range in size from 3.5 to 5.0 kbp, comprising 16.8–17.1 kbp in total [3].
Members of the genus Thelonectria P. Chaverri & Salgado are widespread fungi that belong to the family Nectriaceae Tul. & C. Tul., phylum Ascomycota [4], that can exist in a cylindrocarpon-like asexual state. Typically, they maintain a saprophytic lifestyle; however, they can cause small cankers or root rot on their hosts. Fruiting bodies are mostly spread on the bark of diseased, dying or recently dead broadleaf host trees [5, 6]. To date, no virus has been described infecting a fungus of this genus.
Provenance of the virus material
The Thelonectria strain NW-FVA-1901 (GenBank accession ID: OK161009) was isolated from a necrotic root associated with stem collar necrosis of Fraxinus excelsior L. It was collected in the Waldgehege Fahrenstedthof, mark 24860, Böklund, Abt. 3410a in Schleswig-Holstein, Germany. Isolation and identification as a member of the species Thelonectria was performed as described by Langer [7]. Mycelium was cultivated on potato-dextrose agar (PDA), from which virus-like particles were purified as described by Lutz et al. [8]. Nucleic acids were extracted from particles using a Double-RNA – Viral dsRNA Extraction Kit (iNtRON Biotechnology, Seongnam-Si, South Korea). Isolated dsRNA was subjected to next-generation sequencing. Libraries were prepared using a Nextera XT DNA Library Preparation Kit (Illumina Inc., San Diego, CA, USA) and sequenced on a NextSeq 2000 (Illumina Inc., San Diego, CA, USA) instrument at the Leibniz Institute DSMZ (Braunschweig, Germany) as paired-end reads (2×151). De novo assembly was performed and contigs were analyzed using Geneious Prime software (Biomatters, New Zealand, version 2021.2.2). The 5’ and 3’ termini of each segment were determined by the single-primer amplification technique (SPAT), using an oligonucleotide with a phosphorylated 5’ end and a 2’,3’-dideoxyC group (23ddC) at the 3’ end as a blocker to prevent self-ligation (5’-PO4-TCTCTTCGTGGGCTCTTGCG-23ddC-3’) [9]. Amplicons were cloned into pGEM®-T Vector (Promega Corporation, Madison, Wisconsin, USA) and sequenced. Nucleic acid sequences and ORFs were analyzed using SnapGene Viewer (GSL Biotech, San Diego, CA, USA, version 5.2.4) and BLAST on the NCBI website. Sequence alignments and phylogenetic analysis were performed using MEGA X (version 10.2.4). A bootstrap test was conducted with 1000 replicates for the construction of a maximum-likelihood tree using the Le and Gascuel model with amino acid frequencies and a gamma distribution of 5 (LG+G+F) [10, 11]. Figures were generated and edited using Inkscape (inkscape.org, version 1.1).
Sequence properties
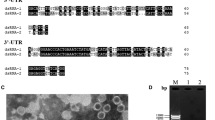
The complete genome sequence of TQV1 has been deposited in the GenBank database (accession ID: OK077750-OK077753). As it is typical for members of the family Quadriviridae [2, 12, 13], each of the four dsRNA segments contains a single ORF on the positive-sense RNA strand (Fig. 2A). The GC content of each segment ranges from 49 to 53%. The sequenced segments corresponded in number and size to the bands detected by agarose gel electrophoresis (Fig. 1), which showed bands ranging from 4.9 to 3.9 kbp.
Agarose gel electrophoresis of dsRNA of TQV1 extracted from isolated virus-like particles from a member of the genus Thelonectria, isolate NW-FVA-1901. M, GeneRuler 1 kb DNA ladder (Thermo Fisher Scientific, Waltham, Massachusetts). Segment 1 to segment 4 of TQV1 range from around 4.9 kbp to 3.9 kbp.
Segment 1 is 4876 bp in length. Its ORF spans from nucleotide position 46 to 4821 and encodes a protein with 1591 aa and a calculated molecular weight of 176.21 kDa. A BLASTp search showed the highest degree of similarity (28.29% aa sequence identity with an E-value of 1e-95) to the hypothetical protein P1 of Botrytis cinerea mycovirus 4 (BcMyV4; accession ID: MN954886.1).
Segment 2 is 4312 bp in length. Its ORF spans from nucleotide position 54 to 4208 and encodes a protein with 1384 aa and a calculated molecular weight of 149.77 kDa. A BLASTp search revealed distant similarities to the structural protein P2 of BcMyV4 (accession ID: MN617035.1; E-value 6e-127; 26.17% aa sequence identity) and the putative capsid protein P2 of Leptosphaeria biglobosa quadrivirus 1 (LbQV-1; accession ID: VCV25422.1; E-value 4e-99; 26.08% aa sequence identity).
Segment 3 is 4158 bp in length. Its ORF spans from nucleotide position 64 to 4119 and encodes a protein with 1351 aa and a calculated molecular weight of 149.00 kDa. A BLASTp search showed similarities to RdRPs of nine confirmed or putative quadriviruses, with an E-value of 0.0. The highest amino acid sequence identity (43.33%) was shared with P3 of Amasya cherry disease-associated mycovirus (ACD; accession ID: CAJ29958.1), and the lowest value (35.28%) was shared with P3 of Rosellinia necatrix quadrivirus 1 (RnQV1; accession ID: BAM93353.1).
Segment 4 is 3933 bp in length. Its ORF spans from nucleotide position 56 to 3418 and encodes a protein with 1120 aa and a calculated molecular weight of 119.98 kDa. A BLASTp search revealed distant similarity (30.00% aa sequence identity) to the hypothetical protein P4 of BcMyV4 (accession ID: QJQ28881.1; E-value 4e-120) [14] as the highest value. The lowest aa sequence identity (26.34%; E-value 3e-19) was shared with the structural protein P4 of RnQV1 (accession ID: YP_005097973.1)
The 5’ UTRs of each segment range in length between 45 bp and 63 bp (dsRNA 1, 45 bp; dsRNA 2, 53 bp; dsRNA 3, 63 bp; dsRNA 4, 55 bp). Segments 2 and 4, which encode putative structural proteins, share 45.6% nt sequence identity at 5’termini of their UTRs. The 3’ UTRs of each segment range from 39 bp to 515 bp (dsRNA 1, 55 bp; dsRNA 2, 104 bp; dsRNA 3, 39 bp; dsRNA 4, 515 bps). All segments possess identical terminal nonamer sequences: 5’-(C/T)ACGAAAAA-3’ at the 5’ terminus and 5’-AT(T/G)AGCAATG(T/C)GC(G/A)CG-3’ at the 3’ terminus. Sequence alignments are shown in Supplementary Fig. S1 and Supplementary Fig. S2, respectively.
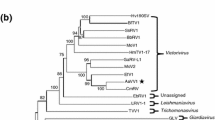
In total, the genome of TQV1 consists of 17.279 bp. To determine the taxonomic position of TQV1, a maximum-likelihood tree was constructed based on a BLASTp search of the sequence of the putative RdRP (P3) of TQV1. It was aligned with P3 sequences of confirmed and tentative members of the families Quadriviridae and Chrysoviridae (Fig. 2B). The phylogenetic analysis showed that TQV1 may represent a new clade between the clade including RnQV1 and the other viruses tentatively assigned to the family Quadriviridae. However, there is currently little sequence information about quadriviruses, and this lack of additional sequences correlates directly with the low bootstrap values observed in the tree. Therefore, the taxonomic position of TQV1 can only be assumed based on this phylogenetic analysis. Nonetheless, based on its genome organization and its phylogenetic position, TQV1 is suggested to be a new member of the family Quadriviridae.
(A) Genome organization of Thelonectria quadrivirus 1 (TQV1). The dsRNA segments are displayed as horizontal lines with their respective UTRs at each terminus. ORFs are represented as boxes with start and stop codon positions indicated above the boxes. Note that the figure is not drawn to scale. (B) Maximum-likelihood tree of TQV1 and selected viruses with 1000 bootstrap replicates. Bootstrap values are displayed at the nodes. The scale bar (0.50) corresponds to the genetic distance. The grey dot indicates the new virus TQV1. The abbreviated names of viruses and dsRNA elements are as follows: CCRS, cherry chlorotic rusty spot associated totiviral-like dsRNA 4; ACD, Amasya cherry disease-associated mycovirus (1294 aa and 1363 aa); BcRV2, Botrytis cinerea RNA virus 2; BcMyV4, Botrytis cinerea mycovirus 4; LbQV-1, Leptosphaeria biglobosa quadrivirus 1; TQV1, Thelonectria quadrivirus 1; RnQV1, Rosellinia necatrix quadrivirus 1; MocV1-D/B, Magnaporthe oryzae chrysovirus 1 D/B
Availability of data and material
The datasets generated and/or analyzed in the current study are available in the GenBank database (accession ID: OK077750-OK077753 and OK161009).
References
Kotta-Loizou I, Coutts RHA (2017) Mycoviruses in Aspergilli: a comprehensive review. Front Microbiol 8:1699. https://doi.org/10.3389/fmicb.2017.01699
Lin Y-H, Chiba S, Tani A, Kondo H, Sasaki A, Kanematsu S et al (2012) A novel quadripartite dsRNA virus isolated from a phytopathogenic filamentous fungus, Rosellinia necatrix. Virology 426:42–50. https://doi.org/10.1016/j.virol.2012.01.013
Chiba S, Castón JR, Ghabrial SA, Suzuki N, Ictv RC (2018) ICTV virus taxonomy profile: Quadriviridae. J Gen Virol 99:1480–1481. https://doi.org/10.1099/jgv.0.001152
Rossman AY, Samuels GJ, Rogerson CT, Lowen R (1999) Genera of Bionectriaceae, Hypocreaceae and Nectriaceae (Hypocreales, Ascomycetes). Stud Mycol 42:1–248
Chaverri P, Salgado C, Hirooka Y, Rossman AY, Samuels GJ (2011) Delimitation of Neonectria and Cylindrocarpon (Nectriaceae, Hypocreales, Ascomycota) and related genera with Cylindrocarpon-like anamorphs. Stud Mycol 68:57–78. https://doi.org/10.3114/sim.2011.68.03
Salgado-Salazar C, Rossman AY, Chaverri P (2016) The genus Thelonectria (Nectriaceae, Hypocreales, Ascomycota) and closely related species with cylindrocarpon-like asexual states. Fungal Divers 80:411–455. https://doi.org/10.1007/s13225-016-0365-x
Langer G (2017) Collar rots in forests of northwest Germany affected by ash dieback. Balt For 23:5–19 (ISSN: 2029-9230)
Lutz T, Petersen JM, Yanık C, de Oliveira C, Heinze C (2021) Processing of the capsid proteins of the betachrysovirus Fusarium graminearum virus-China 9 (FgV-ch9). Virology 563:50–57. https://doi.org/10.1016/j.virol.2021.08.007
Zhong J, Pang XD, Zhu HJ, Da Gao B, Huang WK, Zhou Q (2016) Molecular characterization of a trisegmented mycovirus from the plant pathogenic fungus Colletotrichum gloeosporioides. Viruses 8:268. https://doi.org/10.3390/v8100268
Kumar S, Stecher G, Li M, Knyaz C, Tamura K (2018) MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 35:1547–1549. https://doi.org/10.1093/molbev/msy096
Le SQ, Gascuel O (2008) An improved general amino acid replacement matrix. Mol Biol Evol 25:1307–1320. https://doi.org/10.1093/molbev/msn067
Lin Y-H, Hisano S, Yaegashi H, Kanematsu S, Suzuki N (2013) A second quadrivirus strain from the phytopathogenic filamentous fungus Rosellinia necatrix. Arch Virol 158:1093–1098. https://doi.org/10.1007/s00705-012-1580-8
Shah UA, Kotta-Loizou I, Fitt BDL, Coutts RHA (2018) Identification, molecular characterization, and biology of a novel quadrivirus infecting the phytopathogenic fungus Leptosphaeria biglobosa. Viruses 11:9. https://doi.org/10.3390/v11010009
Ruiz-Padilla A, Rodríguez-Romero J, Gómez-Cid I, Pacifico D, Ayllón MA (2021) Novel mycoviruses discovered in the mycovirome of a necrotrophic fungus. MBio 12:3. https://doi.org/10.1128/mBio.03705-20
Acknowledgements
We thank Birgit Hadeler for technical assistance.
Funding
Open Access funding enabled and organized by Projekt DEAL. This project was financed by the Agency for Renewable Resources (FNR) in the program “Waldklimafonds“ (Forest and Climate Fund) (2219WK22A4 and 2219WK22G4) funded by the German Federal Ministry of Food and Agriculture and the German Federal Ministry for Environment, Nature Conservation and Nuclear Safety.
Author information
Authors and Affiliations
Contributions
Conceptualization: Cornelia Heinze; Methodology: Gitta Langer, Tobias Lutz; Formal analysis and investigation: Tobias Lutz; Writing—original draft preparation: Tobias Lutz; Writing—review and editing: Gitta Langer, Cornelia Heinze, Tobias Lutz; Funding acquisition: Cornelia Heinze, Gitta Langer; Supervision: Cornelia Heinze.
Corresponding author
Ethics declarations
Conflicts of interest
All authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Handling Editor: Massimo Turina.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Lutz, T., Langer, G. & Heinze, C. Complete genome sequence of a new quadrivirus infecting a member of the genus Thelonectria. Arch Virol 167, 691–694 (2022). https://doi.org/10.1007/s00705-021-05353-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-021-05353-y