Abstract
Nanoviruses and geminiviruses are circular, single stranded DNA viruses that infect many plant species around the world. Nanoviruses and certain geminiviruses that belong to the Begomovirus and Mastrevirus genera are associated with additional circular, single stranded DNA molecules (~ 1-1.4 kb) that encode a replication-associated protein (Rep). These Rep-encoding satellite molecules are commonly referred to as alphasatellites and here we communicate the establishment of the family Alphasatellitidae to which these have been assigned. Within the Alphasatellitidae family two subfamilies, Geminialphasatellitinae and Nanoalphasatellitinae, have been established to respectively accommodate the geminivirus- and nanovirus-associated alphasatellites. Whereas the pairwise nucleotide sequence identity distribution of all the known geminialphasatellites (n = 628) displayed a troughs at ~ 70% and 88% pairwise identity, that of the known nanoalphasatellites (n = 54) had a troughs at ~ 67% and ~ 80% pairwise identity. We use these pairwise identity values as thresholds together with phylogenetic analyses to establish four genera and 43 species of geminialphasatellites and seven genera and 19 species of nanoalphasatellites. Furthermore, a divergent alphasatellite associated with coconut foliar decay disease is assigned to a species but not a subfamily as it likely represents a new alphasatellite subfamily that could be established once other closely related molecules are discovered.
Similar content being viewed by others
Introduction
The family Geminiviridae consists of viruses with either monopartite or bipartite circular single-stranded (ss)DNA genomes, with each component individually encapsidated in twinned quasi-isometric (geminate) 22-38 nm virions [3, 17, 55]. The family contains nine genera [46, 48, 54], of which two are relevant to this communication here - Begomovirus and Mastrevirus. Begomoviruses and mastreviruses transmitted by the whitefly Bemisia tabaci and by leafhoppers, respectively, in a circulative manner. The genomes/genomic components of geminiviruses are typically 2600-3600 nucleotides [5, 54].
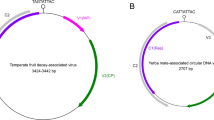
The family Nanoviridae encompasses viruses with multicomponent circular ssDNA genomes that are individually encapsidated within isometric 17-20 nm virions. The family is divided into two genera, Nanovirus and Babuvirus, containing viruses transmitted by aphids in a circulative, non-propagative manner [50, 51]. Members of the genus Nanovirus infect dicotyledonous host plants whereas those of the genus Babuvirus infect monocotyledonous plants. Typically, the genomes of nanoviruses have eight components and the genomes of babuviruses consist of six components. The genomic components of viruses in both genera typically range in size from 970 to 1116 nucleotides [51]. The bona fide genome components of a nanovirus genome share two regions of sequence similarity known as the common region stem-loop (CR-SL) and the common region major (CRM; Figure 1). The DNA-R component of nanoviruses encodes a replication-associated protein (Rep) [7, 12, 13, 15, 16, 25, 38, 42] which is involved in replicating all bona fide genome components.
Illustration of the Rep-encoding DNA molecules of nanovirus- and geminivirus-associated alphasatellites in comparison to the DNA-R components of nanoviruses. The diagrams include the position of the replication-associated protein (Rep), the TATA box of the presumed Rep promoter, an adenine rich sequence (A-rich) in geminivirus-associated alphasatellites and the common region major and common region stem-loop of nanovirus DNA-R components. All components have a predicted stem-loop structure (at position zero) with, in most cases, the nonanucleotide sequence TAGTATTAC forming part of the loop. This likely forms the origin of virion-strand DNA replication that is nicked by Rep to initiate rolling-circle replication
Viruses of the family Nanoviridae, and some begomoviruses and mastreviruses in the family Geminiviridae may be associated with additional circular ssDNA components that resemble the DNA-R components of nanoviruses; these will hereinafter be referred to here as alphasatellites. In common with the DNA-R component of nanoviruses, the alphasatellites encode a Rep protein and have a predicted stem-loop structure with, in most cases, the nonanucleotide sequence TAGTATTAC forming part of the loop (Figure 1). Alphasatellites associated with nanoviruses have a similar size to that of typical nanovirus components (~ 1100 nucleotides), lack the CR-SL and CRM of their helper viruses and are also unable to trans-replicate the bona fide genome components of these helper viruses [18, 43,44,45]. Alphasatellites have also been identified in association with begomoviruses and, in one reported case, a mastrevirus [4, 24]. Typically comprising ~ 1400 nucleotides, the geminivirus associated alphasatellites are significantly larger than both nanovirus components and nanovirus alphasatellites [4]. This size difference, primarily due to the presence in geminivirus alphasatellites of an adenine rich region of sequence and a larger rep gene, is thought to be necessary for the encapsidation of geminivirus alphasatellites by their helper viruses. Geminiviruses have strict restrictions on the size of circular ssDNA molecule that they can encapsidate being half the geminivirus genome length (~ 1400 nucleotides), geminivirus alphasatellites could possibly be encapsidated in isometric (half geminate) virions [8]. For both nanovirus- and geminivirus-associated alphasatellites the sequence relatedness between, and similarity in structure to, nanovirus DNA-R components has led to the hypothesis that the satellites evolved from DNA-R components that were “captured” following co-infections [26, 40].
It is interesting to note that most geminivirus-associated alphasatellites described from the Old World have been associated with monopartite begomoviruses infections that frequently include the presence of betasatellites [39, 52, 56]. In the New World where monopartite begomoviruses are very rare, betasatellites have not been identified and alphasatellites have only ever been reported to occur in association with bipartite Begomovirus infections [10, 32, 37].
The precise impact of alphasatellites on begomovirus and nanovirus infections remains unclear. The presence of nanoalphasatellites in some nanovirus infections has been associated with reduced nanovirus infectivity [45] while geminialphasatellites have been associated with reduced or exacerbated symptoms [27, 32] and/or reduced titer of genomic or betasatellite DNA [19, 21]. Whereas the encoded Rep proteins of alphasatellites associated with the begomoviruses Gossypium darwinii symptomless virus and Gossypium mustelinium symptomless virus suppress post-transcriptional gene silencing [30], those encoded by various other begomovirus alphasatellites are suppressors of transcriptional gene silencing [1]. Due to the rising number of alphasatellites being described, owing mainly to both increasing interest in begomoviruses and nanoviruses, and the ease with which these molecules can be isolated and characterized, there is an urgent need for a robust and workable system of nomenclature and classification for these molecules. Since alphasatellites rely on a helper virus for their spread and are therefore not independent entities, there is very little biological data that can be unequivocally attributed to any of these molecules. It is therefore necessary to base an alphasatellite classification system primarily on information extracted from the nucleotide sequences of these molecules. Such a system could then be adjusted once biological evidence becomes available.
We communicate the creation of the family Alphasatellitidae to which alphasatellites have been assigned and the establishment of two subfamilies based on the fact that there are clear differences between geminivirus- and nanovirus-associated alphasatellites. These include genome length and a distinctive A-rich region downstream of the Rep coding region of the alphasatellites that are associated with geminiviruses. The two sub families are
-
1)
Geminialphasatellitinae: geminivirus-associated alphasatellites
-
2)
Nanoalphasatellitinae: nanovirus-associated alphasatellites
Geminialphasatellitinae
Sequences of geminivirus-associated alphasatellites were downloaded from GenBank (1st May 2017). These were checked to ensure they were complete molecules, had intact Rep open reading frames and were not sub-genomic begomovirus genome components. The filtered 628 geminivirus-associated alphasatellites were analysed with SDT v1.2 [29]. The 628 sequences that remained were aligned using MUSCLE [9] and used to infer a maximum likelihood phylogenetic tree using IQ-TREE [31] with the nucleotide substitution model, GTR+I+G4 (adjudged to be the best fitting by ModelFinder [20]). This tree was rooted with the representative DNA-R sequence of nanoviruses (AB027511, AF102780, AJ290434, AJ749894, EF546807, GU553134, HE654123, JX867550, KC978949, KC979035, KC979054, KX534388, KX534389, KX534390, KX534391, MF535450)
The distribution of the 197506 pairwise identities between each of the possible pair of these 628 sequences had troughs at ~ 70% and ~ 88% pairwise identity (Figure 2) indicating that these values could respectively ben used as genus and species demarcation thresholds, respectively, thus yielding a classification of these sequences with a minimal degree of conflict (Figure 2). These demarcation thresholds are broadly consistent with the viruses in the different species and genera having distinctive helper viruses, geographical distributions and phylogenetic placements (Figure 3; Figure 4).
Distribution of pairwise identities of geminivirus-associated alphasatellites determined using SDT v1.2 [29]
A ‘three color’ pairwise identity matrix inferred using SDT v1.2 [29] showing that both the genera demarcation threshold of 70% and that for species at 88% are supported
Maximum likelihood phylogenetic tree of representative geminivirus-associated alphasatellite sequences from each species inferred using IQ-TREE [31] with GTR+I+G4 chosen as the best-fit model. Branches with less than 60% bootstrap support have been collapsed
The name Alphasatellitidae derives from the name alphasatellite which is now in common usage for this group of satellites. Upon first identification, the begomovirus-associated alphasatellites were called DNA 1 in recognition of their relatedness to the Rep-encoding components of nanoviruses (DNA-R) which at the time were called DNA 1 [26]. The alphasatellite genera names that we have chosen are derived from the names of the type species so, for example, the genus name Colecusatellite derives from the name of the first begomovirus alphasatellite identified, Cotton leaf curl Multan alphasatellite. The alphasatellite species names are, whenever possible, derived from the name of the helper virus with which they were first identified; so cotton leaf curl Multan alphasatellite was first identified in a cotton plant infected with cotton leaf curl Multan virus.
Based on the above criteria, four genera and 43 species are established in the subfamily Geminialphasatellitinae. The classification of the genera and species in the subfamily Geminialphasatellitinae is summarized in Table 1.
The 43 Geminialphasatellitinae species
-
1.
All have a distinctive organization consisting of a single conserved Rep encoding gene in the virion-sense, and a predicted hairpin structure at their presumed origin of virion strand replication that contains, in most cases, a TAGTATTAC loop sequence.
-
2.
With the exception of two alphasatellites, all have been shown to be associated with a geminivirus.
For two alphasatellite sequences that were included in the analysis no helper begomovirus is known. Sequence JX458742 (species Dragonfly associated alphasatellite) was isolated from a dragonfly caught in Puerto Rico and no virus could conclusively be shown to be associated with the molecule. Sequence KT099172 (species Whitefly associated Guatemala alphasatellite 1) was isolated from a B. tabaci whitefly originating from Guatemala and no virus could conclusively be shown to be associated with the molecule.
Guidelines for classification of new geminialphasatellites species
Pairs of complete geminialphasatellite sequences with ≥ 88% pairwise identity calculated using pairwise MUSCLE alignments with similarities calculated ignoring sites with gaps (such as is implemented in SDT v1.2 [29]), should be considered as members of the same species. Those that have < 88% pairwise identity can be considered as new species when coupled with good phylogenetic support.
We recommend that the following steps, similar to those outlined for geminiviruses [6, 28, 46] be taken to resolve such conflicts in cases where 1) a complete geminialphasatellite sequence shares ≥ 88% pairwise identity to sequences that have been assigned to two different species; or 2) a complete geminialphasatellite sequence shares ≥ 88% genome-wide pairwise identity to one or a few sequences assigned to a particular species, even though it shares < 88% identity with the majority of sequences in that species
-
1)
The new geminialphasatellite sequence should be considered as belonging to the species containing sequences with which it shares the highest percentage pairwise identity.
-
2)
The geminialphasatellite should be classified as belonging to any species with which it shares ≥ 88% pairwise identity to any one sequence formerly classified as belonging to that species even if it has < 88% pairwise identity to all other sequences classified as belonging to that species.
Nanoalphasatellitinae
Sequences of nanovirus-associated alphasatellites were downloaded from GenBank (1st May 2017). These sequences were checked to make sure they were complete and had intact Rep open reading frames. The 54 filtered nanovirus-associated alphasatellites were analysed with SDT v1.2 [29]. The sequecnes were aligned using MUSCLE [9] and the resulting alignment was used to infer a maximum likelihood phylogenetic tree using IQ-TREE [31] with the nucleotide substitution model, TIM2+F+G4 (adjudged the best fitting model for these sequences by ModelFinder [20]). The tree was rooted with representative DNA-R sequences of nanoviruses (AB027511, AF102780, AJ290434, AJ749894, EF546807, GU553134, HE654123, JX867550, KC978949, KC979035, KC979054, KX534388, KX534389, KX534390, KX534391, MF535450).
The distribution of 1326 pairwise identities obtained from every possible pair of the 54 nanovirus-associated alphasatellites had troughs at ~ 67% and ~ 80% (Figure 5), and these were chosen respectively as genus and species demarcation cut offs that would yield the minimum number of classification conflicts (Figure 5). The species demarcation threshold implies the existence of at least 19 nanovirus-associated alphasatellite species and the genus demarcation threshold implies the existence of at least seven genera (Figures 6 and 7). The seven genera and 19 species are supported by the phylogenetic clustering of the nanovirus-associated alphasatellite sequences (Figure 7). A summary of the genera and species in the subfamily Nanoalphasatellitinae is provided in Table 1.
Distribution of pairwise identities of nanovirus-associated alphasatellites determined using SDT v1.2 [29]
A ‘three color’ pairwise identity matrix inferred using SDT v1.2 [29] showing that both the genera demarcation threshold of ~ 67% and that for species at 80% are supported
Maximum likelihood phylogenetic trees of the nanovirus-associated alphasatellite sequences inferred using IQ-TREE [31] with TIM2+F+I+G4 chosen as the best fit model. Branches with less than 60% bootstrap support have been collapsed. The Maximum likelihood phylogenetic tree on the left is a guide for genera demarcation with the cyan line showing the rough 67% threshold
Guidelines for classification of new nanoalphasatellites species
Pairs of nanoalphasatellite sequences with ≥ 80% pairwise identity calculated using pairwise MUSCLE alignments with similarities calculated ignoring sites with gaps (such as is implemented in SDT v1.2 [29], should be considered as members of the same species. Those that hare < 80% pairwise identity can be considered as new species when coupled with good phylogenetic support.
We recommend that the following steps, similar to those outlined for geminiviruses [6, 28, 46] be taken to resolve such conflicts in cases where 1) a complete nanoalphasatellite sequence shares ≥ 80% pairwise identity to sequences that have been assigned to two different species; or 2) a complete nanoalphasatellite sequence shares ≥ 80% genome-wide pairwise identity to one or a few sequences assigned to a particular species, even though it shares < 80% identity with the majority of sequences in that species.
-
1)
The new nanoalphasatellite molecule sequence should be considered as belonging to the species containing sequences with which it shares the highest percentage pairwise identity.
-
2)
The nanoalphasatellite should be classified as belonging to any species with which it shares ≥ 80% pairwise identity to any one sequence formerly classified as belonging to that species even if it has < 80% pairwise identity to all other sequences classified as belonging to that species.
Unassigned species in the family Alphasatellitidae
Sequence M29963 (Table 1; species Coconut foliar decay alphasatellite) was isolated from a coconut palm affected by coconut foliar decay disease [36], a disease earlier shown to be associated with a circular ssDNA virus [14, 33,34,35]. However, no virus has so far been identified or characterized infecting coconut even though virions have been described [35]. M29963 does not have all the hallmarks of either Geminialphasatellitinae or Nanoalphasatellitinae alphasatellites in that it has a size (1291 nt) intermediate between the alphasatellites assigned to these sub-families and lacks an A-rich sequence. Furthermore, it was isolated from a monocotyledonous host. Two geminialphasatellites have been isolated from monocotyledonous plants and both of these have features typical of the Geminialphasatellitinae [24]; and twelve have been isolated from bananas and cardamom, all of which have similarities to other nanoalphasatellites [2, 11, 18, 25, 53]. We therefore propose this molecule to represent an unassigned species in the family Alphasatellitidae. It is likely that it is a member of a third sub-family that could be formally proposed once more member sequences have been identified.
Concluding remarks
The last decade has seen a flurry of activity in the identification of circular ssDNA molecules and viruses primarily being driven by multiple displacement amplification techniques coupled with affordable sequencing (both Sanger and high throughput). This has resulted in the identification of a large number of sequences corresponding to diverse viral and viral-like elements sequences from symptomatic and asymptomatic hosts, and from environmental sampling using viral metagenomics approaches. The communication by Simmonds et al. [41] addressed the classification of diverse groups of viral sequences derived from metagenomics approaches and this has resulted in significant changes in viral classification. For example, new families have been established to accommodate virophages and satellite viruses [23] and various novel groups of circular Rep encoding ssDNA viruses derived from environmental sources (with unknown hosts or pathology) [22, 47, 49]. In line with these significant changes on classification of viruses and viral-like elements, we communicate the establishment of a new family Alphasatellitidae with two sub-families Geminialphasatellitinae and Nanoalphasatellitinae for geminivirus- and nanovirus-alphasatellite molecules.
References
Abbas Q, Amin I, Mansoor S, Shafiq M, Wassenegger M, Briddon RW (2017) The Rep proteins encoded by alphasatellites restore expression of a transcriptionally silenced green fluorescent protein transgene in Nicotiana benthamiana. VirusDisease. https://doi.org/10.1007/s13337-017-0413-5
Bell KE, Dale JL, Ha CV, Vu MT, Revill PA (2002) Characterisation of Rep-encoding components associated with banana bunchy top nanovirus in Vietnam. Arch Virol 147:695–707
Bottcher B, Unseld S, Ceulemans H, Russell RB, Jeske H (2004) Geminate structures of African cassava mosaic virus. J Virol 78:6758–6765
Briddon RW, Bull SE, Amin I, Mansoor S, Bedford ID, Rishi N, Siwatch SS, Zafar Y, Abdel-Salam AM, Markham PG (2004) Diversity of DNA 1: a satellite-like molecule associated with monopartite begomovirus-DNA beta complexes. Virology 324:462–474
Brown JK, Fauquet CM, Briddon RW, Zerbini M, Moriones E, Navas-Castillo J (2012) Geminiviridae. In: King AMQ, Adams MJ, Carstens EB, Lefkowitz EJ (eds) Virus taxonomy: ninth report of the International Committee on Taxonomy of Viruse. Elsevier, Amsterdam, pp 351–373
Brown JK, Zerbini FM, Navas-Castillo J, Moriones E, Ramos-Sobrinho R, Silva JC, Fiallo-Olive E, Briddon RW, Hernandez-Zepeda C, Idris A, Malathi VG, Martin DP, Rivera-Bustamante R, Ueda S, Varsani A (2015) Revision of Begomovirus taxonomy based on pairwise sequence comparisons. Arch Virol 160:1593–1619
Burns TM, Harding RM, Dale JL (1995) The genome organization of banana bunchy top virus: analysis of six ssDNA components. J Gen Virol 76:1471–1482
Casado CG, Javier Ortiz G, Padron E, Bean SJ, McKenna R, Agbandje-McKenna M, Boulton MI (2004) Isolation and characterization of subgenomic DNAs encapsidated in “single” T = 1 isometric particles of Maize streak virus. Virology 323:164–171
Edgar RC (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797
Ferro CG, Silva JP, Xavier CAD, Godinho MT, Lima ATM, Mar TB, Lau D, Zerbini FM (2017) The ever increasing diversity of begomoviruses infecting non-cultivated hosts: new species from Sida spp. and Leonurus sibiricus, plus two New World alphasatellites. Ann Appl Biol 170:204–218
Fu HC, Hu JM, Hung TH, Su HJ, Yeh HH (2009) Unusual events involved in Banana bunchy top virus strain evolution. Phytopathology 99:812–822
Gallet R, Kraberger S, Filloux D, Galzi S, Fontes H, Martin DP, Varsani A, Roumagnac P (2018) Nanovirus-alphasatellite complex identified in Vicia cracca in the Rhone delta region of France. Arch Virol 163:695–700
Hafner GJ, Stafford MR, Wolter LC, Harding RM, Dale JL (1997) Nicking and joining activity of banana bunchy top virus replication protein in vitro. J Gen Virol 78:1795–1799
Hanold D, Langridge P, Randles JW (1988) The use of cloned sequences for the Identification of coconut foliar decay disease-associated DNA. J Gen Virol 69:1323–1329
Harding RM, Burns TM, Hafner G, Dietzgen RG, Dale JL (1993) Nucleotide sequence of one component of the banana bunchy top virus genome contains a putative replicase gene. J Gen Virol 74:323–328
Heydarnejad J, Kamali M, Massumi H, Kvarnheden A, Male MF, Kraberger S, Stainton D, Martin DP, Varsani A (2017) Identification of a nanovirus-alphasatellite complex in Sophora alopecuroides. Virus Res 235:24–32
Hipp K, Grimm C, Jeske H, Bottcher B (2017) Near-atomic resolution structure of a plant geminivirus determined by electron cryomicroscopy. Structure 25(1303–1309):e1303
Horser C, Harding R, Dale J (2001) Banana bunchy top nanovirus DNA-1 encodes the ‘master’ replication initiation protein. J Gen Virol 82:459–464
Idris AM, Shahid MS, Briddon RW, Khan AJ, Zhu JK, Brown JK (2011) An unusual alphasatellite associated with monopartite begomoviruses attenuates symptoms and reduces betasatellite accumulation. J Gen Virol 92:706–717
Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS (2017) ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods 14:587–589
Kon T, Rojas MR, Abdourhamane IK, Gilbertson RL (2009) Roles and interactions of begomoviruses and satellite DNAs associated with okra leaf curl disease in Mali, West Africa. J Gen Virol 90:1001–1013
Krupovic M, Ghabrial SA, Jiang D, Varsani A (2016) Genomoviridae: a new family of widespread single-stranded DNA viruses. Arch Virol 161:2633–2643
Krupovic M, Kuhn JH, Fischer MG (2016) A classification system for virophages and satellite viruses. Arch Virol 161:233–247
Kumar J, Kumar J, Singh SP, Tuli R (2014) Association of satellites with a mastrevirus in natural infection: complexity of wheat dwarf India virus disease. J Virol 88:7093–7104
Mandal B, Shilpi S, Barman AR, Mandal S, Varma A (2013) Nine novel DNA components associated with the foorkey disease of large cardamom: evidence of a distinct babuvirus species in Nanoviridae. Virus Res 178:297–305
Mansoor S, Khan SH, Bashir A, Saeed M, Zafar Y, Malik KA, Briddon R, Stanley J, Markham PG (1999) Identification of a novel circular single-stranded DNA associated with cotton leaf curl disease in Pakistan. Virology 259:190–199
Mar TB, Mendes IR, Lau D, Fiallo-Olive E, Navas-Castillo J, Alves MS, Murilo Zerbini F (2017) Interaction between the New World begomovirus Euphorbia yellow mosaic virus and its associated alphasatellite: effects on infection and transmission by the whitefly Bemisia tabaci. J Gen Virol 98:1552–1562
Muhire B, Martin DP, Brown JK, Navas-Castillo J, Moriones E, Zerbini FM, Rivera-Bustamante R, Malathi VG, Briddon RW, Varsani A (2013) A genome-wide pairwise-identity-based proposal for the classification of viruses in the genus Mastrevirus (family Geminiviridae). Arch Virol 158:1411–1424
Muhire BM, Varsani A, Martin DP (2014) SDT: a virus classification tool based on pairwise sequence alignment and identity calculation. PLoS One 9:e108277
Nawaz-Ul-Rehman MS, Nahid N, Mansoor S, Briddon RW, Fauquet CM (2010) Post-transcriptional gene silencing suppressor activity of two non-pathogenic alphasatellites associated with a begomovirus. Virology 405:300–308
Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ (2015) IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol 32:268–274
Paprotka T, Metzler V, Jeske H (2010) The first DNA 1-like alpha satellites in association with New World begomoviruses in natural infections. Virology 404:148–157
Randles JW (1986) Association of single-stranded DNA with the foliar decay disease of coconut palm in Vanuatu. Phytopathology 76:889–894
Randles JW, Hanold D, Julia JF (1987) Small circular single-stranded DNA associated with foliar decay disease of coconut palm in Vanuatu. J Gen Virol 68:273–280
Randles JW, Hanold D (1989) Coconut foliar decay virus particles are 20-nm icosahedra. Intervirology 30:177–180
Rohde W, Randles JW, Langridge P, Hanold D (1990) Nucleotide sequence of a circular single-stranded DNA associated with coconut foliar decay virus. Virology 176:648–651
Romay G, Chirinos D, Geraud-Pouey F, Desbiez C (2010) Association of an atypical alphasatellite with a bipartite New World begomovirus. Arch Virol 155:1843–1847
Sano Y, Wada M, Hashimoto Y, Matsumoto T, Kojima M (1998) Sequences of ten circular ssDNA components associated with the milk vetch dwarf virus genome. J Gen Virol 79:3111–3118
Sattar MN, Kvarnheden A, Saeed M, Briddon RW (2013) Cotton leaf curl disease—an emerging threat to cotton production worldwide. J Gen Virol 94:695–710
Saunders K, Stanley J (1999) A nanovirus-like DNA component associated with yellow vein disease of Ageratum conyzoides: evidence for interfamilial recombination between plant DNA viruses. Virology 264:142–152
Simmonds P, Adams MJ, Benko M, Breitbart M, Brister JR, Carstens EB, Davison AJ, Delwart E, Gorbalenya AE, Harrach B, Hull R, King AM, Koonin EV, Krupovic M, Kuhn JH, Lefkowitz EJ, Nibert ML, Orton R, Roossinck MJ, Sabanadzovic S, Sullivan MB, Suttle CA, Tesh RB, van der Vlugt RA, Varsani A, Zerbini FM (2017) Consensus statement: virus taxonomy in the age of metagenomics. Nat Rev Microbiol 15:161–168
Stainton D, Martin DP, Muhire BM, Lolohea S, Halafihi M, Lepoint P, Blomme G, Crew KS, Sharman M, Kraberger S, Dayaram A, Walters M, Collings DA, Mabvakure B, Lemey P, Harkins GW, Thomas JE, Varsani A (2015) The global distribution of Banana bunchy top virus reveals little evidence for frequent recent, human-mediated long distance dispersal events. Virus Evol 1:vev009
Timchenko T, de Kouchkovsky F, Katul L, David C, Vetten HJ, Gronenborn B (1999) A single rep protein initiates replication of multiple genome components of faba bean necrotic yellows virus, a single-stranded DNA virus of plants. J Virol 73:10173–10182
Timchenko T, Katul L, Sano Y, de Kouchkovsky F, Vetten HJ, Gronenborn B (2000) The master rep concept in nanovirus replication: identification of missing genome components and potential for natural genetic reassortment. Virology 274:189–195
Timchenko T, Katul L, Aronson M, Vega-Arreguin JC, Ramirez BC, Vetten HJ, Gronenborn B (2006) Infectivity of nanovirus DNAs: induction of disease by cloned genome components of Faba bean necrotic yellows virus. J Gen Virol 87:1735–1743
Varsani A, Navas-Castillo J, Moriones E, Hernandez-Zepeda C, Idris A, Brown JK, Murilo Zerbini F, Martin DP (2014) Establishment of three new genera in the family Geminiviridae: Becurtovirus, Eragrovirus and Turncurtovirus. Arch Virol 159:2193–2203
Varsani A, Krupovic M (2017) Sequence-based taxonomic framework for the classification of uncultured single-stranded DNA viruses of the family Genomoviridae. Virus Evol 3:vew037
Varsani A, Roumagnac P, Fuchs M, Navas-Castillo J, Moriones E, Idris A, Briddon RW, Rivera-Bustamante R, Murilo Zerbini F, Martin DP (2017) Capulavirus and Grablovirus: two new genera in the family Geminiviridae. Arch Virol 162:1819–1831
Varsani A, Krupovic M (2018) Smacoviridae: a new family of animal-associated single-stranded DNA viruses. Arch Virol. https://doi.org/10.1007/s00705-018-3820-z
Vetten HJ (2008) Nanoviruses. In: Mahy BWJ, Van Regenmortel M (eds) Encyclopedia of virology. Elsevier, Oxford
Vetten HJ, Dale JL, Grigoras I, Gronenborn B, Harding R, Randles JW, Sano Y, Thomas JE, Timchenko T, Yeh HH (2012) Nanoviridae. In: King AMQ, Adams MJ, Carstens EB, Lefkowitz EJ (eds) Virus taxonomy: ninth report of the International Committee on Taxonomy of Viruse. Elsevier, Amsterdam, pp 395–404
Xie Y, Wu P, Liu P, Gong H, Zhou X (2010) Characterization of alphasatellites associated with monopartite begomovirus/betasatellite complexes in Yunnan, China. Virol J 7:178
Yu NT, Zhang YL, Feng TC, Wang JH, Kulye M, Yang WJ, Lin ZS, Xiong Z, Liu ZX (2012) Cloning and sequence analysis of two banana bunchy top virus genomes in Hainan. Virus Genes 44:488–494
Zerbini FM, Briddon RW, Idris A, Martin DP, Moriones E, Navas-Castillo J, Rivera-Bustamante R, Roumagnac P, Varsani A, Ictv Report C (2017) ICTV virus taxonomy profile: Geminiviridae. J Gen Virol 98:131–133
Zhang W, Olson NH, Baker TS, Faulkner L, Agbandje-McKenna M, Boulton MI, Davies JW, McKenna R (2001) Structure of the Maize streak virus geminate particle. Virology 279:471–477
Zhou X (2013) Advances in understanding begomovirus satellites. Annu Rev Phytopathol 51:357–381
Acknowledgements
The authors are grateful to Drs. J. K. Brown and A. Idris for providing unpublished begomovirus-associated alphasatellite sequences for the preliminary analysis.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare there are no conflicts of interest.
Research involving human participants and/or animals
The research did not involve human participants or animals.
Informed consent
The research did not involve human participants or animals.
Additional information
Handling Editor: Sead Sabanadzovic.
Disclaimer
A.V. is an elected member of the ICTV EC. F.M.Z. is the chair of the Plant Virus Subcommittee of the ICTV. R.W.B., D.P.M., P.R., J.N-C., E.F-O., J-M.L., F.M.Z. and A.V. are current members of the Geminiviridae and Tolecusatellitidae Study Group of the ICTV.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Briddon, R.W., Martin, D.P., Roumagnac, P. et al. Alphasatellitidae: a new family with two subfamilies for the classification of geminivirus- and nanovirus-associated alphasatellites. Arch Virol 163, 2587–2600 (2018). https://doi.org/10.1007/s00705-018-3854-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-018-3854-2