Abstract
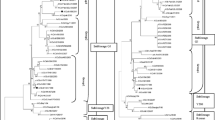
Highly pathogenic avian influenza (HPAI) is a major health concern worldwide. In this study, we focused on antigenic analysis of HPAI H5N1 viruses isolated from poultry in India between 2006 and 2015 comprising 25 isolates from four phylogenetic clades 2.2 (1 isolate), 2.2.2.1 (1 isolate), 2.3.2.1a (17 isolates) and 2.3.2.1c (6 isolates). Seven H5N1 isolates from all four clades were selected for production of chicken antiserum, and antigenic analysis was carried out by hemagglutination inhibition (HI) assay. HI data indicated antigenic divergence (6-21 fold reduction in cross-reactivity) between the two recently emerged clades 2.3.2.1a and 2.3.2.1c. These two clades are highly divergent (21-128 fold reduction in HI titre) from the earlier clades 2.2 /2.2.2.1 isolated in India. However, a maximum of 2-fold and 4-fold reduction in cross-reactivity was observed within the isolates of homologous clades 2.3.2.1c and 2.3.2.1a, respectively. The molecular basis of inter-clade antigenic divergence was examined in the haemagglutinin (HA) antigenic sites of the H5N1 virus. Amino acid changes at 8 HA antigenic sites were observed between clades 2.3.2.1a and 2.3.2.1c, whereas 20-23 substitutions were observed between clades 2.3.2.1a/2.3.2.1c and 2.2/2.2.2.1. Therefore, a systematic analysis of antigenic drift of the contemporary field isolates is a pre-requisite for determining the suitable strain(s) for vaccine candidature.




Similar content being viewed by others
References
Bhat S, Bhatia S, Pillai AS, Sood R, Singh VK, Shrivas OP, Mishra SK, Mawale N (2015) Genetic and antigenic characterization of H5N1 viruses of clade 2.3.2.1 isolated in India. Microb Pathog 88:87–93
Bhatia S, Kunal A, Khandia R, Siddiqui A, Pateriya AK, Sood R (2013) Genetic and antigenic analysis of H5N1 viruses for selection of HA-donor virus for vaccine strains. Indian. J Virol 24:357–364
Bouvier NM, Palese P (2008) The biology of influenza viruses. Vaccine 26(Suppl 4):D49–D53
Cha RM, Smith D, Shepherd E, Davis CT, Donis R, Nguyen T, Nguyen HD, Do HT, Inui K, Suarez DL, Swayne DE, Pantin-Jackwood M (2013) Suboptimal protection against H5N1 highly pathogenic avian influenza viruses from Vietnam in ducks vaccinated with commercial poultry vaccines. Vaccine 31(43):4953–4960
Duvvuri VR, Duvvuri B, Cuff WR, Wu GE, Wu J (2009) Role of positive selection pressure on the evolution of H5N1 hemagglutinin. Genom Proteom Bioinf 7:47–56
Guan Y, Poon LL, Cheung CY, Ellis TM, Lim W, Lipatov AS, Chan KH, Sturm-Ramirez KM, Cheung CL, Leung YH, Yuen KY, Webster RG, Peiris JS (2004) H5N1 influenza: a protean pandemic threat. Proc Natl Acad Sci USA 101(21):8156–8161
Ibrahim M, Eladl AF, Sultan HA, Arafa AS, Razik AGA, Rahman SAE, El-Azm KIA, Saif YM, Lee CW (2013) Antigenic analysis of H5N1 highly pathogenic avian influenza viruses circulating in Egypt (2006–2012). Vet Microbiol 167:651–656
Koel BF, Vliet SV, Burke DF, Bestebroer TM, Bharoto EE, Yasa IWW, Herliana I, Laksono BM, Xu K, Skepner E, Russell CA, Rimmelzwaan GF, Perez DR, Osterhaus ADME, Smith DJ, Prajitno TY, Fouchier RAM (2014) Antigenic variation of clade 2.1 H5N1 virus is determined by a few amino acid substitutions immediately adjacent to the receptor binding site. MBio 5:1–14
Nagarajan S, Tosh C, Smith DK, Peiris JS, Murugkar HV, Sridevi R, Kumar M, Katare M, Jain R, Syed Z, Behera P, Cheung CL, Khandia R, Tripathi S, Guan Y, Dubey SC (2012) Avian Influenza (H5N1) Virus of Clade 2.3.2 in Domestic Poultry in India. PLoS One 7(2):31844
Nguyen T, Rivailler P, Davis CT, Hoa do T, Balish A, Dang NH, Jones J, Vui DT, Simpson N, Huong NT, Shu B, Loughlin R, Ferdinand K, Lindstrom SE, York IA, Klimov A, Donis RO (2012) Evolution of highly pathogenic avian influenza (H5N1) virus populations in Vietnam between 2007 and 2010. Virology 432(2):405–416
OIE (2012) Avian influenza, OIE Terrestrial Manual, Chapter 2.3.4, Paris, France
Pattnaik B, Pateriya AK, Khandia R, Tosh C, Nagarajan S, Gounalan S, Murugkar HV, Shankar BP, Shrivastava N, Behera P, Bhagat S, Peiris JSM, Pradhan HK (2006) Phylogenetic analysis revealed genetic similarity of H5N1 influenza viruses isolated from HPAI outbreaks in chicken in Maharashtra in India with those isolated from Swan in Italy and Iran in 2006. Curr Sci 91:77–81
Pedersen JC (2008) Hemagglutination-inhibition test for avian influenza virus subtype identification and the detection and quantitation of serum antibodies to the avian influenza virus. Methods Mol Biol 436:53–66
Shortridge KF, Zhou NN, Guan Y, Gao P, Ito T, Kawaoka Y, Kodihalli S, Krauss S, Markwell D, Murti KG, Norwood M, Senne D, Sims L, Takada A, Webster RG (1998) Characterization of avian H5N1 influenza viruses from poultry in Hong Kong. Virology 252(2):331–342
Smith DJ, Lapedes AS, de Jong JC, Bestebroer TM, Rimmelzwaan GF, Osterhaus AD, Fouchier RA (2004) Mapping the antigenic and genetic evolution of influenza virus. Science 305(5682):371–376
Tong S, Zhu X, Li Y, Shi M, Zhang J, Bourgeois M, Yang H, Chen X, Recuenco S, Gomez J, Chen LM, Johnson A, Tao Y, Donis RO (2013) New world bats harbor diverse influenza A viruses. PLoS Pathogol 9:1–12
Tosh C, Nagarajan S, Kumar M, Murugkar HV, Venkatesh G, Shukla S, Mishra A, Mishra P, Agarwal S, Singh B, Dubey P, Tripathi S, Kulkarni DD (2016) Multiple introductions of a reassortant H5N1 avian influenza virus of clade 2.3.2.1c with PB2 gene of H9N2 subtype into Indian poultry. Infect Genet Evol 43:173–178
Tosh C, Nagarajan S, Murugkar HV, Bhatia S, Kulkarni DD (2014) Evolution and spread of avian influenza H5N1 viruses. Adv Anim Vet Sci 2:33–41
Tosh C, Nagarajan S, Murugkar HV, Jain R, Behera P, Katare M, Kulkarni DD, Dubey SC (2011) Phylogenetic evidence of multiple introduction of H5N1 virus in Malda district of West Bengal, India in 2008. Vet Microbiol 148:132–139
Webster RG, Bean WJ, Gorman OT, Chambers TM, Kawaoka Y (1992) Evolution and ecology of influenza A viruses. Microbiol Rev 56:152–179
WHO/OIE/FAO H5N1 Evolution Working Group (2012) Continued evolution of highly pathogenic avian influenza A (H5N1): updated nomenclature. Influenza Other Respir Viruses 6:1–5
Acknowledgements
We are thankful to the ICAR, New Delhi and Director, ICAR-NIHSAD, Bhopal for providing necessary facilities to carry out this work. Funding by DADF, MoA, GOI through CDDL grant is acknowledged. S. Bhat acknowledges Director, ICAR-Indian Veterinary Research Institute for financial support.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Funding
This study was supported by CDDL Grant of DADF, Ministry of Agriculture and Farmers Welfare, Government of India. S. Bhat received junior research fellowship from ICAR-Indian Veterinary Research Institute, Izatnagar, Uttar Pradesh.
Conflicts of interest
The authors declare that they have no conflict of interest.
Ethical Standard
The animal experiment was carried out as per the guidelines of CPCSEA, Government of India with approval from Institutional Animal Ethics Committee (IAEC Approval No. 54/IAEC/HSADL/11).
Rights and permissions
About this article
Cite this article
Bhat, S., Nagarajan, S., Kumar, M. et al. Antigenic characterization of H5N1 highly pathogenic avian influenza viruses isolated from poultry in India, 2006-2015. Arch Virol 162, 487–494 (2017). https://doi.org/10.1007/s00705-016-3134-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-016-3134-y