Abstract
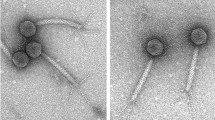
Bacteriophage JBP901, isolated from fermented food, is specific for Bacillus cereus group species and exhibits a broad host spectrum among a large number of B. cereus isolates. Genome sequence analysis revealed a linear 159,492-bp genome with overall G+C content of 39.7 mol%, and 201 ORFs. The presence of a putative methylase, the large number of tRNAs, and the large number of nucleotide-metabolism- and replication-related genes in JBP901 reflects its broad lytic capacity. Most of the ORFs showed a high degree of similarity to Bcp1, Bc431v3 and BCP78, and various comparative genomics analyses also consistently clustered JBP901 with orphan (unclassified) Bacillus phages in the subfamily Spounavirinae of the family Myoviridae, supporting the presence of a distinguishable group in the subfamily.

Similar content being viewed by others

References
Ahmed A (1983) Incidence of Bacillus cereus in milk and some milk products. J Food Prot 46:126–128
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215:403–410
Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, Formsma K, Gerdes S, Glass EM, Kubal M (2008) The RAST Server: rapid annotations using subsystems technology. BMC Genomics 9:75
Bandara N, Kim CH, Kim K-P (2014) Extensive host range determination and improved efficacy of the bacteriophage JBP901 in the presence of divalent cations for control of Bacillus cereus in Cheonggukjang. Food Sci Biotechnol 23:499–504
Bottone EJ (2010) Bacillus cereus, a volatile human pathogen. Clin Microbiol Rev 23:382–398
Delcher AL, Bratke KA, Powers EC, Salzberg SL (2007) Identifying bacterial genes and endosymbiont DNA with Glimmer. Bioinformatics 23:673–679
El-Arabi TF, Griffiths MW, She Y-M, Villegas A, Lingohr EJ, Kropinski AM (2013) Genome sequence and analysis of a broad-host range lytic bacteriophage that infects the Bacillus cereus group. Virol J 10:48
Granum PE, Lund T (1997) Bacillus cereus and its food poisoning toxins. FEMS Microbiol Lett 157:223–228
Guinebretière M-H, Broussolle V (2002) Enterotoxigenic profiles of food-poisoning and food-borne Bacillus cereus strains. J Clin Microbiol 40:3053–3056
Hagens S, Loessner MJ (2010) Bacteriophage for biocontrol of foodborne pathogens: calculations and considerations. Curr Pharm Biotechnol 11:58–68
Hugas M, Tsigarida E, Robinson T, Calistri P (2007) Risk assessment of biological hazards in the European Union. Int J Food Microbiol 120:131–135
Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C (2012) Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28:1647–1649
Ki Kim S, Kim K-P, Jang SS, Mi Shin E, Kim M-J, Oh S, Ryu S (2009) Prevalence and toxigenic profiles of Bacillus cereus isolated from dried red peppers, rice, and Sunsik in Korea. J Food Prot 72:578–582
King AMQ, Adams MJ, Lefkowitz EJ, Carstens EB (2012) Virus taxonomy: classification and nomenclature of viruses: ninth report of the International Committee on Taxonomy of Viruses. Elsevier Academic Press
Klumpp J, Lavigne R, Loessner MJ, Ackermann H-W (2010) The SPO1-related bacteriophages. Arch Virol 155:1547–1561
Lavigne R, Darius P, Summer EJ, Seto D, Mahadevan P, Nilsson AS, Ackermann HW, Kropinski AM (2009) Classification of Myoviridae bacteriophages using protein sequence similarity. BMC Microbiol 9:224
Lee G-I, Lee H-M, Lee C-H (2012) Food safety issues in industrialization of traditional Korean foods. Food Cont 24:1–5
Lee J-H, Shin H, Son B, Ryu S (2012) Complete genome sequence of Bacillus cereus bacteriophage BCP78. J Virol 86:637–638
Lowe TM, Eddy SR (1997) tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res 25:0955–0964
Lukashin AV, Borodovsky M (1998) GeneMark. hmm: new solutions for gene finding. Nucleic Acids Res 26:1107–1115
Mahadevan P, King JF, Seto D (2009) CGUG: in silico proteome and genome parsing tool for the determination of “core” and unique genes in the analysis of genomes up to ca. 1.9 Mb. BMC Res Notes 2:168
Oguntoyinbo FA, Mathew Oni O (2004) Incidence and characterization of Bacillus cereus isolated from traditional fermented meals in Nigeria. J Food Prot 67:2805–2808
Park Y-B, Kim J-B, Shin S-W, Kim J-C, Cho S-H, Lee B-K, Ahn J, Kim J-M, Oh D-H (2009) Prevalence, genetic diversity, and antibiotic susceptibility of Bacillus cereus strains isolated from rice and cereals collected in Korea. J Food Prot 72:612–617
Sambrook J, Russell DW, Russell DW (2001) Molecular cloning: a laboratory manual (3-volume set). Cold Spring Harbor Laboratory Press, Cold Spring Harbor
Schuch R, Pelzek AJ, Fazzini MM, Nelson DC, Fischetti VA (2014) Complete genome sequence of Bacillus cereus sensu lato bacteriophage Bcp1. Genome Announc 2:e00334-00314
Shin H, Bandara N, Shin E, Ryu S, Kim KP (2011) Prevalence of Bacillus cereus bacteriophages in fermented foods and characterization of phage JBP901. Res Microbiol 162:791–797
Solovyev V, Salamov A (2011) Automatic annotation of microbial genomes and metagenomic sequences. Metagenomics and its applications in agriculture, biomedicine and environmental studies. Nova Science Publishers, Hauppauge, pp 61–78
Stewart CR, Casjens SR, Cresawn SG, Houtz JM, Smith AL, Ford ME, Peebles CL, Hatfull GF, Hendrix RW, Huang WM (2009) The genome of Bacillus subtilis bacteriophage SPO1. J Mol Biol 388:48–70
Sullivan MJ, Petty NK, Beatson SA (2011) Easyfig: a genome comparison visualizer. Bioinformatics 27:1009–1010
Thorsen L, Azokpota P, Hansen BM, Hounhouigan DJ, Jakobsen M (2010) Identification, genetic diversity and cereulide producing ability of Bacillus cereus group strains isolated from Beninese traditional fermented food condiments. Int J Food Microbiol 142:247–250
Thorsen L, Azokpota P, Munk Hansen B, Rønsbo MH, Nielsen KF, Hounhouigan DJ, Jakobsen M (2011) Formation of cereulide and enterotoxins by Bacillus cereus in fermented African locust beans. Food Microbiol 28:1441–1447
Van der Auwera GA, Feldgarden M, Kolter R, Mahillon J (2013) Whole-genome sequences of 94 environmental isolates of Bacillus cereus sensu lato. Genome Announc 1:e00380-00313
Wong HC, Chang M, Fan J (1988) Incidence and characterization of Bacillus cereus isolates contaminating dairy products. Appl Environ Microbiol 54:699–702
Yee LM, Matsumoto T, Yano K, Matsuoka S, Sadaie Y, Yoshikawa H, Asai K (2011) The genome of Bacillus subtilis phage SP10: a comparative analysis with phage. Biosci Biotechnol Biochem 75:944–952
Acknowledgments
This research was supported by High Value-Added Food Technology Development Program (project No. 112108-2), Korea Institute of Planning and Evaluation for Technology in Food, Agriculture, Forestry and Fisheries (IPET) and the Ministry for Food, Agriculture, Forestry, and Fisheries of Republic of Korea, and by the “Cooperative Research Program for Agriculture & Technology Development (Project No. PJ009842)”, Rural Development Administration, Republic of Korea.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Asare, P.T., Ryu, S. & Kim, KP. Complete genome sequence and phylogenetic position of the Bacillus cereus group phage JBP901. Arch Virol 160, 2381–2384 (2015). https://doi.org/10.1007/s00705-015-2485-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-015-2485-0