Abstract
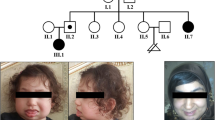
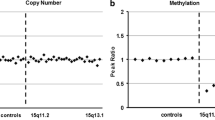
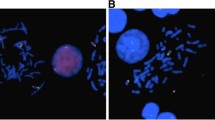
X-chromosome inactivation (XCI) is an essential mechanism in females that compensates for the genome imbalance between females and males. It is known that XCI can spread into an autosome of patients with X;autosome translocations. The subject was a 5-year-old boy with Prader–Willi syndrome (PWS)-like features including hypotonia, hypo-genitalism, hypo-pigmentation, and developmental delay. G-banding, fluorescent in situ hybridization, BrdU-incorporated replication, human androgen receptor gene locus assay, SNP microarrays, ChIP-on-chip assay, bisulfite sequencing, and real-time RT-PCR were performed. Cytogenetic analyses revealed that the karyotype was 46,XY,der(X)t(X;15)(p21.1;q11.2),−15. In the derivative chromosome, the X and half of the chromosome 15 segments showed late replication. The X segment was maternal, and the chromosome 15 region was paternal, indicating its post-zygotic origin. The two chromosome 15s had a biparental origin. The DNA methylation level was relatively high in the region proximal from the breakpoint, and the level decreased toward the middle of the chromosome 15 region; however, scattered areas of hypermethylation were found in the distal region. The promoter regions of the imprinted SNRPN and the non-imprinted OCA2 genes were completely and half methylated, respectively. However, no methylation was found in the adjacent imprinted gene UBE3A, which contained a lower density of LINE1 repeats. Our findings suggest that XCI spread into the paternal chromosome 15 led to the aberrant hypermethylation of SNRPN and OCA2 and their decreased expression, which contributes to the PWS-like features and hypo-pigmentation of the patient. To our knowledge, this is the first chromosome-wide methylation study in which the DNA methylation level is demonstrated in an autosome subject to XCI.






Similar content being viewed by others
References
Akahoshi K, Fukai K, Kato A, Kimiya S, Kubota T, Spritz RA (2001) Duplication of 15q11.2-q14, including the P gene, in a woman with generalized skin hyperpigmentation. Am J Med Genet 104:299–302
Bailey JA, Carrel L, Chakravarti A, Eichler EE (2000) Molecular evidence for a relationship between LINE-1 elements and X chromosome inactivation: the Lyon repeat hypothesis. Proc Natl Acad Sci USA 97:6634–6639
Brilliant MH, King R, Francke U, Schuffenhauer S, Meitinger T, Gardner JM, Durham-Pierre D, Nakatsu Y (1994) The mouse pink-eyed dilution gene: association with hypopigmentation in Prader–Willi and Angelman syndromes and with human OCA2. Pigment Cell Res 7:398–402
Brown CJ, Ballabio A, Rupert JL, Lafreniere RG, Grompe M, Tonlorenzi R, Willard HF (1991) A gene from the region of the human X inactivation centre is expressed exclusively from the inactive X chromosome. Nature 349:38–44
Chow JC, Ciaudo C, Fazzari MJ, Mise N, Servant N, Glass JL, Attreed M, Avner P, Wutz A, Barillot E, Greally JM, Voinnet O, Heard E (2010) LINE-1 activity in facultative heterochromatin formation during X chromosome inactivation. Cell 141:956–969
DuBose AJ, Johnstone KA, Smith EY, Hallett RA, Resnick JL (2010) Atp10a, a gene adjacent to the PWS/AS gene cluster, is not imprinted in mouse and is insensitive to the PWS-IC. Neurogenetics 11:145–151
Gartler SM, Riggs AD (1983) Mammalian X-chromosome inactivation. Annu Rev Genet 17:155–190
Giorda R, Bonaglia MC, Milani G, Baroncini A, Spada F, Beri S, Menozzi G, Rusconi M, Zuffardi O (2008) Molecular and cytogenetic analysis of the spreading of X inactivation in a girl with microcephaly, mild dysmorphic features and t(X;5)(q22.1;q31.1). Eur J Hum Genet 16:897–905
Herzing LB, Romer JT, Horn JM, Ashworth A (1997) Xist has properties of the X-chromosome inactivation centre. Nature 386:272–275
Ke X, Collins A (2003) CpG islands in human X-inactivation. Ann Hum Genet 67:242–249
Kubota T, Nonoyama S, Tonoki H, Masuno M, Imaizumi K, Kojima M, Wakui K, Shimadzu M, Fukushima Y (1999) A new assay for the analysis of X-chromosome inactivation based on methylation-specific PCR. Hum Genet 104:49–55
Kubota T, Wakui K, Nakamura T, Ohashi H, Watanabe Y, Yoshino M, Kida T, Okamoto N, Matsumura M, Muroya K, Ogata T, Goto Y, Fukushima Y (2002) The proportion of cells with functional X disomy is associated with the severity of mental retardation in mosaic ring X Turner syndrome females. Cytogenet Genome Res 99:276–284
Lee JT, Jaenisch R (1997) Long-range cis effects of ectopic X-inactivation centres on a mouse autosome. Nature 386:275–279
Lee JT, Strauss WM, Dausman JA, Jaenisch R (1996) A 450 kb transgene displays properties of the mammalian X-inactivation center. Cell 86:83–94
Lyon MF (1961) Gene action in the X-chromosome of the mouse (Mus musculus L.). Nature 190:372–373
Lyon MF (1962) Sex chromatin and gene action in the mammalian X-chromosome. Am J Hum Genet 14:135–148
Lyon MF (1998) X-chromosome inactivation: a repeat hypothesis. Cytogenet Cell Genet 80:133–137
Manola KN, Stavropoulou C, Georgakakos VN, Zoi K, Fisfis M, Evmorfiadis I, Zoi C, Pantelias GE, Stefanoudaki K, Sambani C (2007) Switch in X-inactivation in a JAK2 V617F-negative case of polycythemia vera with two acquired X-autosome translocations. Leuk Res 31:1009–1014
Ogata T, Wakui K, Muroya K, Ohashi H, Matsuo N, Brown DM, Ishii T, Fukushima Y (1998) Microphthalmia with linear skin defects syndrome in a mosaic female infant with monosomy for the Xp22 region: molecular analysis of the Xp22 breakpoint and the X-inactivation pattern. Hum Genet 103:51–56
Orellana C, Martinez F, Badia L, Millan JM, Montero MR, Andres J, Prieto F (2001) Trisomy rescue by postzygotic unbalanced (X;14) translocation in a girl with dysmorphic features. Clin Genet 60:206–211
Popova BC, Tada T, Takagi N, Brockdorff N, Nesterova TB (2006) Attenuated spread of X-inactivation in an X;autosome translocation. Proc Natl Acad Sci USA 103:7706–7711
Schmidt M, Du Sart D (1992) Functional disomies of the X chromosome influence the cell selection and hence the X inactivation pattern in females with balanced X-autosome translocations: a review of 122 cases. Am J Med Genet 42:161–169
Sharp A, Robinson DO, Jacobs P (2001) Absence of correlation between late-replication and spreading of X inactivation in an X;autosome translocation. Hum Genet 109:295–302
Sharp AJ, Spotswood HT, Robinson DO, Turner BM, Jacobs PA (2002) Molecular and cytogenetic analysis of the spreading of X inactivation in X;autosome translocations. Hum Mol Genet 11:3145–3156
Sharp AJ, Migliavacca E, Dupre Y, Stathaki E, Sailani MR, Baumer A, Schinzel A, Mackay DJ, Robinson DO, Cobellis G, Cobellis L, Brunner HG, Steiner B, Antonarakis SE (2010) Methylation profiling in individuals with uniparental disomy identifies novel differentially methylated regions on chromosome 15. Genome Res 20:1271–1278
Solari AJ, Rahn IM, Ferreyra ME, Carballo MA (2001) The behavior of sex chromosomes in two human X-autosome translocations: failure of extensive X-inactivation spreading. Biocell 25:155–166
Stankiewicz P, Kuechler A, Eller CD, Sahoo T, Baldermann C, Lieser U, Hesse M, Glaser C, Hagemann M, Yatsenko SA, Liehr T, Horsthemke B, Claussen U, Marahrens Y, Lupski JR, Hansmann I (2006) Minimal phenotype in a girl with trisomy 15q due to t(X;15)(q22.3;q11.2) translocation. Am J Med Genet A 140:442–452
Takagi N, Abe K (1990) Detrimental effects of two active X chromosomes on early mouse development. Development 109:189–201
Uehara S, Hanew K, Harada N, Yamamori S, Nata M, Niikawa N, Okamura K (2001) Isochromosome consisting of terminal short arm and proximal long arm X in a girl with short stature. Am J Med Genet 99:196–199
Vassiliou GS, Campbell PJ, Li J, Roberts I, Swanton S, Huntly BJ, Fourouclas N, Baxter EJ, Munro LR, Culligan DA, Scott LM, Green AR (2006) An acquired translocation in JAK2 Val617Phe-negative essential thrombocythemia associated with autosomal spread of X-inactivation. Haematologica 91:1100–1104
Wang Z, Willard HF, Mukherjee S, Furey TS (2006) Evidence of influence of genomic DNA sequence on human X chromosome inactivation. PLoS Comput Biol 2:e113
White WM, Willard HF, Van Dyke DL, Wolff DJ (1998) The spreading of X inactivation into autosomal material of an x;autosome translocation: evidence for a difference between autosomal and X-chromosomal DNA. Am J Hum Genet 63:20–28
Wirth J, Back E, Huttenhofer A, Nothwang HG, Lich C, Gross S, Menzel C, Schinzel A, Kioschis P, Tommerup N, Ropers HH, Horsthemke B, Buiting K (2001) A translocation breakpoint cluster disrupts the newly defined 3′ end of the SNURF-SNRPN transcription unit on chromosome 15. Hum Mol Genet 10:201–210
Xue F, Tian XC, Du F, Kubota C, Taneja M, Dinnyes A, Dai Y, Levine H, Pereira LV, Yang X (2002) Aberrant patterns of X chromosome inactivation in bovine clones. Nat Genet 31:216–220
Yang X, Smith SL, Tian XC, Lewin HA, Renard JP, Wakayama T (2007) Nuclear reprogramming of cloned embryos and its implications for therapeutic cloning. Nat Genet 39:295–302
Acknowledgments
We thank the patient and their parents for their cooperation in this study and Professor Emeritus Tadashi Kajii for his critical reading of the manuscript. This research was supported in part by Grants-in-Aid for Scientific Research (S.S. and T.K.), Exploratory Research (T.K.) from the Ministry of Education, Culture Sports, Science, and Technology, Japan, and the Kawano foundation for medical research (T.K.), and Kawano Masanori Foundation for Promotion of Pediatrics (S.S.).
Conflict of interest
None.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Sakazume, S., Ohashi, H., Sasaki, Y. et al. Spread of X-chromosome inactivation into chromosome 15 is associated with Prader–Willi syndrome phenotype in a boy with a t(X;15)(p21.1;q11.2) translocation. Hum Genet 131, 121–130 (2012). https://doi.org/10.1007/s00439-011-1051-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00439-011-1051-4