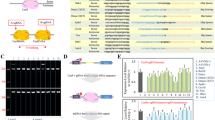
Abstract
Previous works using human tripronuclear zygotes suggested that the clustered regularly interspaced short palindromic repeat (CRISPR)/Cas9 system could be a tool in correcting disease-causing mutations. However, whether this system was applicable in normal human (dual pronuclear, 2PN) zygotes was unclear. Here we demonstrate that CRISPR/Cas9 is also effective as a gene-editing tool in human 2PN zygotes. By injection of Cas9 protein complexed with the appropriate sgRNAs and homology donors into one-cell human embryos, we demonstrated efficient homologous recombination-mediated correction of point mutations in HBB and G6PD. However, our results also reveal limitations of this correction procedure and highlight the need for further research.




Similar content being viewed by others
References
Capmany G, Taylor A, Braude PR, Bolton VN (1996) The timing of pronuclear formation, DNA synthesis and cleavage in the human 1-cell embryo. Mol Hum Reprod 2:299–306
Chapman JR, Taylor MR, Boulton SJ (2012) Playing the end game: DNA double-strand break repair pathway choice. Mol Cell 47:497–510
Chen F, Pruett-Miller SM, Huang Y, Gjoka M, Duda K, Taunton J, Collingwood TN, Frodin M, Davis GD (2011) High-frequency genome editing using ssDNA oligonucleotides with zinc-finger nucleases. Nat Methods 8:753–755
Chen S, Lee B, Lee AY, Modzelewski AJ, He L (2016) Highly efficient mouse genome editing by CRISPR ribonucleoprotein electroporation of zygotes. J Biol Chem 291:14457–14467
Cho SW, Lee J, Carroll D, Kim JS, Lee J (2013) Heritable gene knockout in Caenorhabditis elegans by direct injection of Cas9-sgRNA ribonucleoproteins. Genetics 195:1177–1180
Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, Zhang F (2013) Multiplex genome engineering using CRISPR/Cas systems. Science 339:819–823
Du CS, Ren X, Chen L, Jiang W, He Y, Yang M (1999) Detection of the most common G6PD gene mutations in Chinese using amplification refractory mutation system. Hum Hered 49:133–138
Hashimoto M, Yamashita Y, Takemoto T (2016) Electroporation of Cas9 protein/sgRNA into early pronuclear zygotes generates non-mosaic mutants in the mouse. Dev Biol 418:1–9
Hasty P, Rivera-Perez J, Bradley A (1991) The length of homology required for gene targeting in embryonic stem cells. Mol Cell Biol 11:5586–5591
Heyer WD, Ehmsen KT, Liu J (2010) Regulation of homologous recombination in eukaryotes. Annu Rev Genet 44:113–139
Kang X, He W, Huang Y, Yu Q, Chen Y, Gao X, Sun X, Fan Y (2016) Introducing precise genetic modifications into human 3PN embryos by CRISPR/Cas-mediated genome editing. J Assist Reprod Genet 33:581–588
Kim S, Kim D, Cho SW, Kim J, Kim JS (2014) Highly efficient RNA-guided genome editing in human cells via delivery of purified Cas9 ribonucleoproteins. Genome Res 24:1012–1019
Kimura A, Matsunaga E, Takihara Y, Nakamura T, Takagi Y, Lin S, Lee H (1983) Structural analysis of a beta-thalassemia gene found in Taiwan. J Biol Chem 258:2748–2749
Lee JS, Kwak SJ, Kim J, Kim AK, Noh HM, Kim JS, Yu K (2014) RNA-guided genome editing in Drosophila with the purified Cas9 protein. G3 (Bethesda) 4:1291–1295
Li W, Teng F, Li T, Zhou Q (2013) Simultaneous generation and germline transmission of multiple gene mutations in rat using CRISPR-Cas systems. Nat Biotechnol 31:684–686
Liang P, Xu Y, Zhang X, Ding C, Huang R, Zhang Z, Lv J, Xie X, Chen Y, Li Y, Sun Y, Bai Y, Songyang Z, Ma W, Zhou C, Huang J (2015) CRISPR/Cas9-mediated gene editing in human tripronuclear zygotes. Protein Cell 6:363–372
Lieber MR (2010) The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu Rev Biochem 79:181–211
Lin S, Staahl BT, Alla RK, Doudna JA (2014a) Enhanced homology-directed human genome engineering by controlled timing of CRISPR/Cas9 delivery. Elife 3:e04766
Lin Y, Cradick TJ, Brown MT, Deshmukh H, Ranjan P, Sarode N, Wile BM, Vertino PM, Stewart FJ, Bao G (2014b) CRISPR/Cas9 systems have off-target activity with insertions or deletions between target DNA and guide RNA sequences. Nucleic Acids Res 42:7473–7485
Ma Y, Shen B, Zhang X, Lu Y, Chen W, Ma J, Huang X, Zhang L (2014) Heritable multiplex genetic engineering in rats using CRISPR/Cas9. PLoS One 9:e89413
Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM (2013) RNA-guided human genome engineering via Cas9. Science 339:823–826
Niu Y, Shen B, Cui Y, Chen Y, Wang J, Wang L, Kang Y, Zhao X, Si W, Li W, Xiang AP, Zhou J, Guo X, Bi Y, Si C, Hu B, Dong G, Wang H, Zhou Z, Li T, Tan T, Pu X, Wang F, Ji S, Zhou Q, Huang X, Ji W, Sha J (2014) Generation of gene-modified cynomolgus monkey via Cas9/RNA-mediated gene targeting in one-cell embryos. Cell 156:836–843
Ran FA, Hsu PD, Lin CY, Gootenberg JS, Konermann S, Trevino AE, Scott DA, Inoue A, Matoba S, Zhang Y, Zhang F (2013) Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell 154:1380–1389
Shen B, Zhang J, Wu H, Wang J, Ma K, Li Z, Zhang X, Zhang P, Huang X (2013) Generation of gene-modified mice via Cas9/RNA-mediated gene targeting. Cell Res 23:720–723
Wang H, Yang H, Shivalila CS, Dawlaty MM, Cheng AW, Zhang F, Jaenisch R (2013) One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell 153:910–918
Zhou J, Shen B, Zhang W, Wang J, Yang J, Chen L, Zhang N, Zhu K, Xu J, Hu B, Leng Q, Huang X (2014) One-step generation of different immunodeficient mice with multiple gene modifications by CRISPR/Cas9 mediated genome engineering. Int J Biochem Cell Biol 46:49–55
Acknowledgements
We thank Dr. Yuanfeng Li of Beijing Proteome Research Center and Dr. Zhongsheng Sun of Beijing Institutes of Life Science, Chinese Academy of Sciences for help with genome sequencing and analysis. We thank Dr. Xingxu Huang of the School of Life Science and Technology, Shanghai Tech University for helpful advice.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Funding
This study was funded by a Grant from the Beijing Municipal Committee on Science and Technology (#Z141100000214015), by an international collaboration Grant from the Chinese Minister of Science and Technology (#2013DFB30210), by The National Basic Research Program (973 Program, No. 2013CB910300), by a Grant from the Science and Technology Program of Guangzhou, China (#2014KZDXM047), by The Innovation of Science and Technology Commission of Guangzhou, China (No. 201604020075), by The Department of Science and Technology of Guangdong, China (No. 2016A020218012), and a Grant from the China Postdoctoral Science Foundation (#2013M532214).
Conflict of interest
All authors declare that they have no conflict of interest.
Ethical approval
This study was approved by the ethics committee of the Third Affiliated Hospital of Guangzhou Medical University, numbered [2015] No. 068. The methods used in the present study closely followed the guidelines legislated and posted by the Ministry of Health of the People’s Republic of China. The patients involved in this study knew about and understood the usage of tripronuclear zygotes, immature oocytes and leftover sperm, and voluntarily donated them after providing informed consent.
Informed consent
Informed consent was obtained from all individual participants included in the study.
Additional information
Communicated by S. Hohmann.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Tang, L., Zeng, Y., Du, H. et al. CRISPR/Cas9-mediated gene editing in human zygotes using Cas9 protein. Mol Genet Genomics 292, 525–533 (2017). https://doi.org/10.1007/s00438-017-1299-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00438-017-1299-z