Abstract
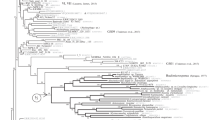
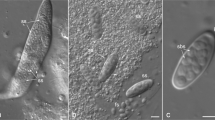
Microsporidia are intracellular eukaryotic parasites of animals, characterized by unusual morphological and genetic features. They can be divided in three main groups, the classical microsporidians presenting all the features of the phylum and two putative primitive groups, the chytridiopsids and metchnikovellids. Microsporidia originated from microsporidia-like organisms belonging to a lineage of chytrid-like endoparasites basal or sister to the Fungi. Genetic and genomic data are available for all members, except chytridiopsids. Herein, we filled this gap by obtaining the rDNA sequence (SSU-ITS-partial LSU) of Chytridiopsis typographi (Chytridiopsida), a parasite of bark beetles. Our rDNA molecular phylogenies indicate that Chytridiopsis branches earlier than metchnikovellids, commonly thought ancestral, forming the more basal lineage of the Microsporidia. Furthermore, our structural analyses showed that only classical microsporidians present 16S-like SSU rRNA and 5.8S/LSU rRNA gene fusion, whereas the standard eukaryote rRNA gene structure, although slightly reduced, is still preserved in the primitive microsporidians, including 18S-like SSU rRNA with conserved core helices, and ITS2-like separating 5.8S from LSU. Overall, our results are consistent with the scenario of an evolution from microsporidia-like rozellids to microsporidians, however suggesting for metchnikovellids a derived position, probably related to marine transition and adaptation to hyperparasitism. The genetic and genomic data of additional members of Chytridiopsida and Rozellomycota will be of great value, not only to resolve phylogenetic relationships but also to improve our understanding of the evolution of these fascinating organisms.





Similar content being viewed by others
References
Bass D, Czech L, Williams BAP, Berney C, Dunthorn M, Mahé F, Torruella G, Stentiford GD, Williams TA (2018) Clarifying the relationships between microsporidia and Cryptomycota. J Eukaryot Microbiol. https://doi.org/10.1111/jeu.12519
Beard CB, Butler JF, Becnel JJ (1990) Nolleria pulicis n. gen., n. sp. (microsporidia: Chytridiopsidae), a microsporidian parasite of the cat flea, Ctenocephalides felis (Siphonaptera: Pulicidae). J Protozool 37(2):90–99. https://doi.org/10.1111/j.1550-7408.1990.tb05876.x
Blackwell WH, Letcher PM, Powell MJ (2016) Reconsideration of the inclusiveness of genus Plasmophagus (Chytridiomycota, posteris traditus) based on morphology. Phytologia 98(2):128–136
Blackwell WH, Letcher PM, Powell MJ (2017) The taxa of Dictyomorpha (Chytridiomycota, in praesens tempus). Phytologia 99(1):77–82
Burke JM (1970) A microsporidian in the epidermis of Eisenia foetida (Oligochaeta). J Invertebr Pathol 16(1):145–147. https://doi.org/10.1016/0022-2011(70)90222-3
Canning EU, Refardt D, Vossbrinck CR, Okamura B, Curry A (2002) New diplokaryotic microsporidia (phylum microsporidia) from freshwater bryozoans (Bryozoa, Phylactolaemata). Eur J Protistol 38(3):247–266. https://doi.org/10.1078/0932-4739-00867
Coleman AW (2007) Pan-eukaryote ITS-2 homologies revealed by RNA secondary structure. Nucleic Acids Res 35(10):3322–3329. https://doi.org/10.1093/nar/gkm233
Corradi N (2015) Microsporidia: eukaryotic intracellular parasites shaped by gene loss and horizontal gene transfers. Annu Rev Microbiol 69:167–183. https://doi.org/10.1146/annurev-micro-091014-104136
Corsaro D, Walochnik J, Venditti D, Steinmann J, Müller KD, Michel R (2014a) Microsporidia-like parasites of amoebae belong to the early fungal lineage Rozellomycota. Parasitol Res 113(5):1909–1918. https://doi.org/10.1007/s00436-014-3838-4
Corsaro D, Walochnik J, Venditti D, Müller K-D, Hauröder B, Michel R (2014b) Rediscovery of Nucleophaga amoebae, a novel member of the Rozellomycota. Parasitol Res 113(12):4491–4498. https://doi.org/10.1007/s00436-014-4138-8
Corsaro D, Michel R, Walochnik J, Venditti D, Müller KD, Hauröder B, Wylezich C (2016) Molecular identification of Nucleophaga terricolae sp. nov. (Rozellomycota), and new insights on the origin of the microsporidia. Parasitol Res 115(8):3003–3011. https://doi.org/10.1007/s00436-016-5055-9
Cuomo CA, Desjardins CA, Bakowski MA, Goldberg J, Ma AT, Becnel JJ, Didier ES, Fan L, Heiman DI, Levin JZ, Young S, Zeng Q, Troemel ER (2012) Microsporidian genome analysis reveals evolutionary strategies for obligate intracellular growth. Genome Res 22(12):2478–2488. https://doi.org/10.1101/gr.142802.112
Dangeard P-A (1895) Mémoire sur les parasites du noyau et du protoplasme. Le Botaniste 4:199–248
Dawson SC, Pace NR (2002) Novel kingdom-level eukaryotic diversity in anoxic environments. Proc Natl Acad Sci U S A 99(12):8324–8329 www.pnas.org/cgi/doi/10.1073/pnas.062169599
De Puytorac P, Tourret M (1963) Étude de kystes d’origine parasitaire (Microsporidies ou Grégarines) sur le parois interne du corps des vers Megascolecidae. Ann Parasitol (Paris) 38(6):861–874
De Rijk P, Wuyts, De Wachter R (2003) RnaViz 2: an improved representation of RNA secondary structure. Bioinformatics 19(2):299–300
Franzen C (2004) Microsporidia: how can they invade other cells. Trends Parasitol 20(6):275–279. https://doi.org/10.1016/j.pt.2004.04.009
Grossart HP, Wurzbacher C, James TY, Kagami M (2016) Discovery of dark matter fungi in aquatic ecosystems demands a reappraisal of the phylogeny and ecology of zoosporic fungi. Fungal Ecol 19:28–38. https://doi.org/10.1016/j.funeco.2015.06.004
Haag KL, James TY, Pombert JF, Larsson R, Schaer TM, Refardt D, Ebert D (2014) Evolution of a morphological novelty occurred before genome compaction in a lineage of extreme parasites. Proc Natl Acad Sci U S A 111(43):15480–15485. https://doi.org/10.1073/pnas.1410442111
Han B, Weiss LM (2017) Microsporidia: obligate intracellular pathogens within the fungal kingdom. Microbiol Spectrum 5(2):FUNK-0018-2016. https://doi.org/10.1128/microbiolspec.FUNK-0018-2016
James TY, Pelin A, Bonen L, Ahrendt S, Sain D, Corradi N, Stajich JE (2013) Shared signatures of parasitism and phylogenomics unite Cryptomycota and microsporidia. Curr Biol 23(16):1548–1553. https://doi.org/10.1016/j.cub.2013.06.057
Jobb G, von Haeseler A, Strimmer K (2004) TREEFINDER: a powerful graphical analysis environment for molecular phylogenetics. BMC Evol Biol 4(18):18. https://doi.org/10.1186/1471-2148-4-18
Katinka MD, Duprat S, Cornillot E, Méténier G, Thomarat F, Prensier G, Barbe V, Peyretaillade E, Brottier P, Wincker P, Delbac F, El Alaoui H, Peyret P, Saurin W, Gouy M, Weissenbach J, Vivarès CP (2001) Genome sequence and gene compaction of the eukaryote parasite Encephalitozoon cuniculi. Nature 414(6862):450–453. https://doi.org/10.1038/35106579
Keeling PJ, Corradi N, Morrison HG, Haag KL, Ebert D, Weiss LM, Akiyoshi DE, Tzipori S (2010) The reduced genome of the parasitic microsporidian Enterocytozoon bieneusi lacks genes for core carbon metabolism. Genome Biol Evol 2:304–309. https://doi.org/10.1093/gbe/evq022
Lanave C, Preparata G, Saccone C, Serio G (1984) A new method for calculating evolutionary substitution rates. J Mol Evol 20(1):86–93. https://doi.org/10.1007/BF02101990
Larsson JIR (1980) Insect pathological investigations on Swedish Thysanura. II. A new microsporidian parasite of Petrobius brevistylis (Microcoryphia, Machilidae): description of the species and creation of two new genera and a new family. Protistologica 16:85–101
Larsson (1993) Description of Chytridiopsis trichopterae N. Sp. (Microspora, Chytridiopsidae), a microsporidian parasite of the caddis fly Polycentropus flavomaculatus (Trichoptera, Polycentropodidae), with comments on relationships between the families Chytridiopsidae and Metchnikovellidae. J Eukaryot Microbiol 40(1):37–48. https://doi.org/10.1111/j.1550-7408.1993.tb04880.x
Larsson JIR (2000) The hyperparasitic microsporidium Amphiacantha longa Caullery et Mesnil, 1914 (Microspora: Metchnikovellidae) - description of the cytology, redescription of the species, emended diagnosis of the genus Amphiacantha and establishment of the new family Amphiacanthidae. Folia Parasitol 47(4):241–256
Larsson JIR (2014) The primitive microsporidia. In: Weiss LM, Becnel JJ (eds) Microsporidia: pathogens of opportunity. Wiley, Chichester, pp 605–634
Larsson JIR, Køie M (2006) The ultrastructure and reproduction of Amphiamblys capitellides (Microspora, Metchnikovellidae), a parasite of the gregarine Ancora sagittata (Apicomplexa, Lecudinidae), with redescription of the species and comments on the taxonomy. Eur J Protistol 42(4):233–248. https://doi.org/10.1016/j.ejop.2006.07.001
Larsson JIR, Steiner MY, Bjørnson S (1997) Intexta acarivora gen. Et sp. n. (Microspora: Chytridiopsidae) – ultrastructural study and description of a new microsporidian parasite of the forage mite Tyrophagus putrescentiae (Acari: acaridae). Acta Protozool 36(4):295–304
Lubinsky G (1955) On some parasites of parasitic protozoa: II. Sagittospora cameroni gen. n., sp. n.—a phycomycete parasitizing Ophryoscolecidae. Can J Microbiol 1(8):675–684. https://doi.org/10.1139/m55-081
Manier J-F, Ormieres R (1968) Ultrastructure de quelques stades de Chytridiopsis socius Schn., parasite de Blaps lethifera Marsh. (Coleopt., Tenebr.). Protistologica 4:181–185
Mikhailov KV, Simdyanov TG, Aleoshin VV (2016) Genomic survey of a hyperparasitic microsporidian Amphiamblys sp. (Metchnikovellidae). Genome Biol Evol 9(3):454–467. https://doi.org/10.1093/gbe/evw235
Morris DJ, Terry RS, Ferguson KB, Smith JE, Adams A (2005) Ultrastructural and molecular characterization of Bacillidium vesiculoformis n. sp. (Microspora: Mrazekiidae) in the freshwater oligochaete Nais simplex (Oligochaeta: Naididae). Parasitology 130(1):31–40
Purrini K, Weiser J (1985) Ultrastructural study of the microsporidian Chytridiopsis typographi (Chytridiopsida: Microspora) infecting the bark beetle Ips typographus (Scolytidae:Coleoptera), with new data on spore dimorphism. J Invertebr Pathol 45(1):66–74. https://doi.org/10.1016/0022-2011(85)90051-5
Quandt CA, Beaudet D, Corsaro D, Walochnik J, Michel R, Corradi N, James TY (2017) The genome of an intranuclear parasite, Paramicrosporidium saccamoebae, reveals alternative adaptations to obligate intracellular parasitism. eLife 6:e29594. https://doi.org/10.7554/eLife.29594.
Radek R, Kariton M, Dabert J, Alberti G (2015) Ultrastructural characterization of Acarispora falculifera n.gen., n.sp., a new microsporidium (Opisthokonta: Chytridiopsida) from the feather mite Falculifer rostratus (Astigmata: Pterolichoidea). Acta Parasitol 60(2):200–210. https://doi.org/10.1515/ap-2015-0029
Ronquist F, Huelsenbeck JP (2003) MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19(12):1572–1574. https://doi.org/10.1093/bioinformatics/btg180
Sokolova YY, Kryukova NA, Glupov VV, Fuxa JR (2006) Systenostrema alba Larsson 1988 (microsporidia, Thelohaniidae) in the dragonfly Aeshna viridis (Odonata, Aeshnidae) from South Siberia: morphology and molecular characterization. J Eukaryot Microbiol 53(1):49–57. https://doi.org/10.1111/j.1550-7408.2005.00075.x
Sokolova YY, Paskerova GG, Rorati YM, Nassonova ES, Smirnov AV (2013) Fine structure of Metchnikovella incurvata Caullery and Mesnil 1914 (microsporidia), a hyperparasite of gregarines Polyrhabdina sp. from the polychaete Pygospio elegans. Parasitology 140(7):855–867. https://doi.org/10.1017/S0031182013000036
Sokolova YY, Paskerova GG, Rotari YM, Nassonova ES, Smirnov AV (2014) Description of Metchnikovella spiralis sp. n. (microsporidia: Metchnikovellidae), with notes on the ultrastructure of metchnikovellids. Parasitology 141(8):1108–1122. https://doi.org/10.1017/S0031182014000420
Sparrow FK Jr (1960) Aquatic Phycomycetes, 2nd edn. The University of Michigan Press, Ann Arbor
Sprague V (1977) Classification and phylogeny of the microsporidia. In: Bulla LA, Cheng TC (eds) Comparative pathobiology, Systematics of the microsporidia, vol 2. Plenum Press, New York, pp 1–30
Stentiford GD, Ramilo A, Abollo E, Kerr R, Bateman KS, Feist SW, Bass D, Villalba A (2017) Hyperspora aquatica n. gn., n. sp. (microsporidia), hyperparasitic in Marteilia cochillia (Paramyxida), is closely related to crustacean-infecting microspordian taxa. Parasitology 144(2):186–199. https://doi.org/10.1017/S0031182016001633
Thélohan P (1894) Sur la présence d’une capsule à filament dans les spores microsporidies. C R Acad Ac Paris 118:1425–1427
Tsaousis AD, Kunji ERS, Goldberg AV, Lucocq JM, Hirt RP, Embley TM (2008) A novel route for ATP acquisition by the remnant mitochondria of Encephalitozoon cuniculi. Nature 453(7194):553–556. https://doi.org/10.1038/nature06903
Vaughn JC, Sperbeck S, Ramsey WJ, Lawrence CB (1984) A universal model for the secondary structure of 5.8S ribosomal RNA molecules, their contact sites with 28S ribosomal RNAs, and their prokaryotic equivalent. Nucleic Acids Res 12(19):7479–7502
Vávra J, Lukeš J (2013) Microsporidia and ‘the art of living together’. Adv Parasitol 82:253–319. https://doi.org/10.1016/B978-0-12-407706-5.00004-6
Vivier E (1965) Étude, au microscope électronique, de la spore de Metchnikovella hovassei n. sp.: appartenance des Metchnikovellidae aux Microsporidies. C R Seances Soc Biol Paris 260:6982–6984
Wegensteiner R, Weiser J (1996) Occurrence of Chytridiopsis typographi (Microspora, Chytridiopsida) in Ips typographus L. (Coleoptera, Scolytidae) field populations and in a laboratory stock. J Appl Entomol 120:595–602. https://doi.org/10.1111/j.1439-0418.1996.tb01657.x
Wegensteiner R, Weiser J, Führer E (1996) Observations on the occurrence of pathogens in the bark beetle Ips typographus L. (Col.,Scolytidae). J Appl Entomol 120:190–204. https://doi.org/10.1111/j.1439-0418.1996.tb01591.x
Wegensteiner R, Tkaczuk C, Bałazy S, Griesser S, Rouffaud MA, Stradner A, Steinwender BM, Hager H, Papierok B (2015) Occurrence of pathogens in populations of Ips typographus, Ips sexdentatus (Coleoptera, Curculionidae, Scolytinae) and Hylobius spp. (Coleoptera, Curculionidae, Curculioninae) from Austria, Poland and France. Acta Protozool 54(3):219–232. https://doi.org/10.4467/16890027AP.15.018.3215
Weiser J (1977) Contribution of the classification of microsporidia. Acta Soc Zool Bohem 41(4):308–320
Williams BAP, Hirt RP, Lucocq JM, Embley TM (2002) A mitochondrial remnant in the microsporidian Trachipleistophora hominis. Nature 418(6900):865–869. https://doi.org/10.1038/nature00949
Wuyts J, Van de Peer Y, De Wachter R (2001) Distribution of substitution rates and location of insertion sites in the tertiary structure of ribosomal RNA. Nucleic Acids Res 29(24):5017–5028
Xu Y, Weiss LM (2005) The microsporidian polar tube: a highly specialised invasion organelle. Int J Parasitol 35(9):941–953. https://doi.org/10.1016/j.ijpara.2005.04.003
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Section Editor: Yaoyu Feng
Electronic supplementary material
Suppl. Fig. 1
Secondary structure of the SSU rRNA of Amphiacantha sp. (ex Lecudina cf. elongata). The sequence was retrieved from GenBank (ID KX214676). Both the 5′- and 3′-ends are lacking. (PDF 791 kb)
Suppl. Fig. 2
Secondary structure of the SSU rRNA of Amphiamblys sp. WSBS2006 (ex Ancora sagittata). The sequence was retrieved from GenBank (ID KX214672). (PDF 167 kb)
Rights and permissions
About this article
Cite this article
Corsaro, D., Wylezich, C., Venditti, D. et al. Filling gaps in the microsporidian tree: rDNA phylogeny of Chytridiopsis typographi (Microsporidia: Chytridiopsida). Parasitol Res 118, 169–180 (2019). https://doi.org/10.1007/s00436-018-6130-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00436-018-6130-1