Abstract
Purpose
Dysregulation of miRNA profile has been associated with a broad spectrum of cellular processes underlying progression of various human malignancies. Increasing evidence suggests that specific microRNA clusters might be of clinical utility, especially in triple-negative breast carcinoma (TNBC), devoid of both predictive markers and potential therapeutic targets. Here we provide a comprehensive review of the existing data on microRNAs in TNBC, their molecular targets, a putative role in invasive progression with a particular emphasis on the epithelial-to-mesenchymal transition (EMT) and acquisition of stem-cell properties (CSC), regarded both as prerequisites for metastasis, and significance for therapy.
Methods
PubMed and Medline databases were systematically searched for the relevant literature. 121 articles have been selected and thoroughly analysed.
Results
Several miRNAs associated with EMT/CSC and invasion were identified as significantly (1) upregulated: miR-10b, miR-21, miR-29, miR-9, miR-221/222, miR-373 or (2) downregulated: miR-145, miR-199a-5p, miR-200 family, miR-203, miR-205 in TNBC. Dysregulation of miR-10b, miR-21, miR-29, miR-145, miR-200 family, miR-203, miR-221/222 was reported of prognostic value in TNBC patients.
Conclusion
Available data suggest that specific microRNA clusters might play an important role in biology of TNBC, understanding of which should assist disease prognostication and therapy.
Similar content being viewed by others
Introduction
Despite continuous advances in early detection and development of personalized therapy, breast cancer (BCa) is still the leading cause of death from cancer among women, with age-standardized mortality rate of 12.9/100,000 worldwide (Ferlay et al. 2014; Tao et al. 2015). Comprehensive gene expression profiling has identified five major BCa molecular subtypes (luminal A, luminal B, HER2-type, triple-negative and normal-like BCa) characterised by specific morphological patterns and distinct biological properties and, more importantly, different clinical course and prognosis (Perou et al. 2000; Polyak 2007). The most aggressive, triple-negative breast cancer (TNBC), lacks expression of valid predictive markers [oestrogen receptor (ER), progesterone receptor (PR) and epidermal growth factor receptor 2 (HER2)], and thus devoid of clear therapeutic targets, it presents a serious clinical challenge. Patients with TNBC do not benefit from endocrine or HER2-targeted therapies and have worse outcome after chemotherapies in comparison to sufferers from other BCa subtypes (Lehmann and Pietenpol 2015). Shortened disease-free and overall survival of TNBC patients calls for urgent identification of new molecular targets that may improve prognostication and, above all, assist in development of efficient specific therapies.
MicroRNAs (miRNAs) are small, non-coding, endogenous, universal RNA regulators of key biological processes (Lin and Gregory 2015). In cancer, dysregulation of miRNA profile has been associated with mechanisms of disease development, including activation of invasiveness and metastasis (Lin and Gregory 2015). In TNBC, in particular, emerging in vitro and in vivo studies identified various miRNAs likely to be linked to the aggressive phenotype (Chang et al. 2015; Zhu et al. 2017; Lü et al. 2017; Paszek et al. 2017; Avery-Kiejda et al. 2017).
As metastatic process is considered the major cause of cancer-related death, our review focuses on key microRNAs of potential clinical value in TNBC, involved in regulation of main mechanisms underlying invasive progression, particularly, the epithelial-to-mesenchymal transition (EMT) and acquisition of stem cell-like properties (CSC).
Triple-negative breast cancer—molecular features
Systemic investigation of gene expression patterns in human breast tumours revealed the molecular taxonomy of BCa dividing them into five subgroups dependent on genetic and biological similarities (Perou et al. 2000). The microarray analyses identified the triple-negative breast cancer (ER-, PR- and HER2-negative) as a clinically heterogeneous malignancy and the most aggressive BCa subtype that is characterised by high rates of tumour recurrence and poor overall survival. Aggressive phenotype of TNBC defined by poor disease-free survival, high recurrence rate and shortened time of overall survival is connected with biological and clinical factors, including high nuclear grade, high histological grade, high genomic instability, loss of suppressor genes, as well as gain of migratory, invasive and stem cell-like properties of cancer cells (Arpino et al. 2015).
MicroRNAs
Biology of miRNAs
MicroRNAs comprise a class of 22–25 nucleotides long, non-coding, endogenous RNA molecules, which play important regulatory roles by targeting mRNA transcripts, leading to their translational repression or degradation (Lin and Gregory 2015). Biogenesis of miRNA is under tight spatial and temporal control and is cell- and tissue-specific (Lin and Gregory 2015). In human, majority of miRNAs are encoded among introns, however, their presence was also observed in exonic regions. Production of miRNAs starts when so-called pri-miRNAs are transcribed by RNA polymerase II/III. Clustered pri-miRNAs are further converted into pre-miRNAs by a microprocessor complex consisting of RNAse III enzyme Drosha and DGCR8 (DiGeorge Critical Region 8) protein (Lee et al. 2002, 2003). Drosha cleaves 11 bp away from the single-stranded stem loop junction, converting pri-miRNA into pre-miRNA, which contains 5′ phosphate group and 2–3 nucleotides 3′ overhang (Lee et al. 2002, 2003). Pre-miRNA is translocated to the cytoplasm and cleaved by a specific endonuclease—RNAse III Dicer to finally form a single-stranded, mature miRNA (Hutvágner et al. 2001). This is then activated by the Argonaute family protein and coupled into the microRNA-induced silencing complex (miRISC), which attaches to the 3′ untranslated region (3′ UTR) of the target mRNA (Fire et al. 1998; Hannon et al. 2000; Martinez et al. 2002). The degree of complementarity between miRNA and its target mRNA determines efficacy of miRNA action. While a perfect match leads to mRNA deadenylation, and consequently its degradation, an imperfect pairing only inhibits translation of the target mRNA.
Regulatory role of miRNAs
MicroRNAs regulate diverse cellular processes (Hwang and Mendell 2006; Shivdasani 2006; Olivieri et al. 2013), and thus create a characteristic signature/profile reflecting both tissue-specificity and developmental stage (differentiation) of the cell. For example, expression changes of specific miRNA clusters are highly informative and may be used to identify the tissue of origin of poorly differentiated tumour (Hwang and Mendell 2006). In TNBC, various miRNAs have been associated with processes essential to disease progression, such as epithelial-to-mesenchymal transition (EMT), acquisition of stem-like properties by cancer cells, migration, invasiveness, and metastatic spread.
Epithelial-to-mesenchymal transition
EMT, regarded as a prerequisite for metastasis, is a cellular reprogramming mechanism crucial to the ‘invasive makeover’ of cancer cells (Sethi et al. 2011; Seton-Rogers 2016; Felipe Lima et al. 2016). A fundamental event in EMT, marking the onset of the process, is the loss of E-cadherin expression, which in normal epithelial cells is required for maintenance of integrity of the entire cadherin–catenin–actin network. Regulation of E-cadherin expression at the transcriptional level is not fully understood yet, but several transcription factors, such as SNAI1/Snail1, SLUG, ZEB1, ZEB2, E47 and KLF8 (Kruppel-like factor 8) have been shown to bind to the E-cadherin promoter and repress directly its transcription (Singh and Settleman 2010; Lamouille et al. 2014; Seton-Rogers 2016; Felipe Lima et al. 2016).
The epithelial–mesenchymal switch involves changes in several pathways, including TGF-β, WNT, HIF1/2, NOTCH, NF-κB and RAS-ERK1/2. In the TGF-β pathway, the signal is generated from the TGF-β-activated kinase receptors (TGF-βRI and TGF-βRII) and processed downwards by the SMAD-dependent (formation of the activated complex of SMAD2, SMAD3 and SMAD4) or SMAD-independent (including PI3K/AKT and ERK/MAPK) signalling. This results in overexpression of SNAIL1/2 and ZEB1/2 and, finally, in the repression of E-cadherin expression (Shirakihara et al. 2011; Massagué 2012; Weiss and Attisano 2013). Activation of the WNT pathway, found particular significance in breast cancer, results in the stabilization of β-catenin, leading to the subsequent break of its interaction with E-cadherin and translocation into nucleus, where it participates in the induction of the mesenchymal-specific gene profile (Yook et al. 2006; Niehrs 2012; Lamouille et al. 2014). Notch signalling induces EMT both directly and via the crosstalk with other signalling pathways. Direct signals created by binding of Delta or Jagged family ligands to the Notch receptors cause cleavage of Notch intracellular domain (NICD), which migrates to the nucleus, where it promotes expression of SNAIL1/2 (Kaidi et al. 2007; Sahlgren et al. 2008; Wu et al. 2009; Espinoza et al. 2013). In Hedgehog (Hh) signalling, glioma 1-associated protein (GLI1), the Hh effector, promotes SNAIL1/2 expression (Kalluri and Weinberg 2009; Thiery et al. 2009). EMT can also be triggered by the signals induced by growth factor (FGF, EGF, HGF, VEGF) receptors involved in activation of the RAS-ERK1/2 or PI3K-Akt pathways. The EMT process is also strongly influenced by stimuli derived from tumour microenvironment, such as inflammation, hypoxia and metabolic or oncogenic stress (Kalluri and Weinberg 2009; Yuan et al. 2012). The existence of a positive feedback loop between pro-inflammatory microenvironment and EMT has been shown in several experiments (Mantovani et al. 2008; López-Novoa and Nieto 2009). Cohen et al. reported that inflammatory cytokines induced EMT in inflammatory breast cancer cell lines (SUM149PT, KPL4, IBC-3, SUM190PT), whereas their inhibitors blocked the process (Cohen et al. 2015). Stimulation of normal breast epithelial cells with inflammatory cytokines, such as tumour necrosis factor alpha (TNF-α) and interleukin 1beta (Il-1β), resulted in induction of EMT (mainly by upregulation of ZEB-1 and SNAIL expression), and this was associated with acquirement of invasive phenotype by the cells (Leibovich-Rivkin et al. 2013) Reversely, overexpression of the key EMT transcription factors led to increased secretion of pro-inflammatory cytokines IL-1, IL-6, IL8 by the cells, further stimulating EMT (Ricciardi et al. 2015). Decrease of reactive oxygen species caused by tumour-related hypoxia upregulated hypoxia-inducible factors (HIF1/2), which led to EMT via overexpression of ZEB1/2 and protection of SNAIL from degradation (Wang and Zhou 2011; Lamouille et al. 2013).
MiRNA-200 family, which embraces miR-200a, miR-200b, miR-200c, miR-141 and miR-429, are well-known negative regulators of EMT, specifically targeting ZEB1/2 (Table 1; Fig. 1) (Korpal et al. 2008; Kalluri and Weinberg 2009; Wang et al. 2013, 2014; Humphries et al. 2014; Zaravinos and Apostolos 2015). Functional in vitro studies identified miR-200 family as downregulated in TNBC cells and confirmed their tumour-suppressive action in normal tissues (Korpal et al. 2008; Mekala et al. 2018). Humphries and colleagues showed significantly lower levels of miR-200 family members in metastatic TNBCs in comparison to other subtypes of breast cancer (Humphries et al. 2014). MiRNA-200 family contributes to the pathogenesis of TNBC via various pathways, including BRCA1/2, however, their most significant impact is exerted via regulation of EMT (Korpal et al. 2008; Humphries et al. 2014; Erturk et al. 2015). The interdependence between miRs-200 family and EMT was reported by Korpal and colleagues, who in NMuMG cells (murine mammary epithelial cells—a model of TGFβ1-induced EMT) observed a strong downregulation of all miR-200 family members (except for miR-141) upon stimulation with TGF (Korpal et al. 2008). Moreover, overexpression of miR-200 resulted in EMT repression in NMuMG cells. MiR-200 was linked to an increase of E-cadherin level and reversal of mesenchymal phenotype in 4TO7 cells, the murine TNBC cell line (Korpal et al. 2008). Gregory and co-workers obtained consistent results and found that miR-200 level was markedly lower in cells undergone EMT in response to the TGFβ treatment (Gregory et al. 2008) and inhibition of miR-200 was sufficient for induction of EMT via upregulation of ZEB1. Decreased expression of miR-200 family was detected in mesenchymal-like TNBC invasive human BCa cell lines (MDA-MB-435, BT-549) (Gregory et al. 2008). Overexpression of miR-200c in TNBC cells (MDA-MB-231 cell line) resulted in loss of the elongated shape associated with a motile, mesenchymal phenotype and acquisition of the epithelial-like morphology. Downregulation of miR-200b was found crucial in increase of EMT in TNBC cells by targeting ZEB1/2 and suppressing PKCα (Kolacinska et al. 2014; Humphries et al. 2014; Rhodes et al. 2015). Loss of the actin-based structure was orchestrated by miR-200c, which directly targeted actin regulatory proteins, FHOD1 and PPM1F, in a ZEB1/2-independent manner and led to the inhibition of migration and invasion of the cells (Jurmeister et al. 2012).
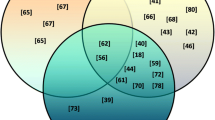
Schematic presentation of microRNAs involvement in regulation of epithelial-to mesenchymal transition (EMT) and acquisition of stem cell-like properties (CSC) in triple-negative breast carcinoma. Red/green arrows—upregulated/downregulated microRNAs; black arrows/truncated lines—activation (upregulation)/repression (inhibition) of signalling pathways
MiR-205, in addition to the miR-200 family, has been revealed by microarray analyses as significantly reduced in cells induced to undergo EMT (Gregory et al. 2008; Piovan et al. 2012). MiR-205 expression in mesenchymal-like BCa cells is strongly downregulated in comparison to that in cells with the epithelial phenotype. Interestingly, microRNA expression profiling has shown decreased expression of miR-205 in TNBC, suggestive of its tumour-suppressive role (Huo et al. 2016). Gregory et al. demonstrated that in MDA-MB-231, MDA-MB-435 and BT-549 cells, miR-205 suppressed ZEB1/2 and, reversely, induction of EMT via TGFβ led to decrease of miR-205 (Gregory et al. 2008). This reciprocal regulatory loop between miR-205 and ZEB1/2 transcription factors is similar to that described for the miR-200 family (Gregory et al. 2008; Chao et al. 2014).
MiR-199a-5p is another example of microRNA that confers tumour-suppressive role and is downregulated in TNBC. Ectopic expression of miR-199a-5p in MDA-MB-231 cells significantly altered expression pattern of EMT-related genes such as CDH1, ZEB1 and TWIST, leading to the inhibition of the process. Moreover, Chen et al. demonstrated that elevated level of miR-199a-5p impaired cell motility and invasiveness as well as inhibited tumour growth in vivo (Chen et al. 2016).
MiR-9 represents a group of microRNAs upregulated in TNBC (Table 1). MiR-9-mediated downregulation of E-cadherin leads to activation of β-catenin signalling pathway and upregulation of VEGF expression. In TNBC, miR-9 was shown to be associated with MYC amplification, tumour grade and metastatic status (Ma et al. 2010; Jang et al. 2017). High level of miR-9 correlated with poor disease-free survival (DFS) and distant metastasis-free survival (DMSF) (Ma et al. 2010). D’Ippolito et al. observed higher expression of miR-9 in TNBC in comparison to the luminal and HER2-enriched BCa subtypes. Moreover, upon ligand activation of PDGFRβ signalling, miR-9 promoted formation of vascular-like structures by TNBC cells both in vitro and in vivo, whereas inhibition of miR-9 expression strongly decreased the number of vascular lacunae (D’Ippolito et al. 2016). Interestingly, miR-9 may also act as an important player in the cross-talk between tumour and its stroma, as exosome-mediated delivery of miR-9 was shown to induce cancer-associated fibroblast properties in normal human mammary fibroblasts (Baroni et al. 2016).
MiR-221/222, reported to be overexpressed in TNBC, is involved in yet another mechanism of EMT regulation (Table 1) (Stinson et al. 2011). As demonstrated by Stinson et al., miR-221 and its direct target, TRSP1, repress ZEB2 expression leading to E-cadherin downregulation (Stinson et al. 2011). Depletion of adiponectin receptor 1 (ADIPOR1), another miR-221’s target, induced EMT in MCF10A cells, activated NFκB and JAK2/STAT3 signalling pathways as well as increased cell migration and invasion (Hwang et al. 2013). Knockdown of miR-221 blocked cell cycle progression, promoted cell apoptosis and inhibited in vitro proliferation and tumour in vivo growth. Silencing of miR-221 increased also expression of E-cadherin, and decreased SLUG and Snail level in TNBC cell lines, such as MDA-MB-231, BT-20, and MDA-MB-468 (Nassirpour et al. 2013).
Cancer stem cells (CSC)
Reactivation of the stem cell programme is a phenomenon closely associated with sustained cancer progression as well as failure of eradicating therapies. By generating pools of self-renewing cancer stem cells (CSCs), primary and metastatic tumours, especially of the most aggressive type as TNBC, become more resistant to chemo- and radiotherapy (Scheel and Weinberg 2012; Hollier et al. 2013; Lim et al. 2013; Wang et al. 2015). The interdependence between EMT and breast cancer CSCs (BCSCs) was shown in various in vitro and animal models, confirming a well-documented direct link between the EMT and acquisition of stem cell-like properties (Shostak and Chariot 2011; Yamamoto et al. 2013; D’Angelo et al. 2015; Jang et al. 2015). Many signalling pathways implied in the induction of EMT, such as Wnt, Notch, TNFα, NFκB or TGFβ, control also CSCs functions (Shostak and Chariot 2011; Yamamoto et al. 2013; D’Angelo et al. 2015; Jang et al. 2015). Similarly, microRNAs involved in regulation of EMT, contribute to induction and maintenance of stemness as well as influence CSCs response to the EMT-related signals (Fig. 1).
MiR-203 is one of the well described microRNAs, which are involved in both stemness and EMT in TNBC. In normal breast epithelial cells, miR-203 is correlated with cell differentiation by targeting ΔNp63α, the predominant TP63 isoform in mammary epithelia, vital to the maintenance of epithelial stemness. Overexpression of miR-203 induced reversal of EMT, the mesenchymal-to-epithelial transition (MET), and led to decreased proliferation and colony formation of MDA-MB-231 cells (DeCastro et al. 2013). Moreover, Wang et al. observed that upregulation of miR-203 in TNBC cell lines (MDA-MB-468, MDA-MB-231) resulted in growth and invasion inhibition, enhancement of cell differentiation and reduction of cell metastatic capacity (DeCastro et al. 2013; Zhao et al. 2015). Taube et al. showed that miR-203 was repressed via epigenetic modification (DNA methylation) to a greater degree in TNBC cells (MDA-MB-231, SUM-159) than in more differentiated luminal BCa cell lines (MCF7, T47D) (Zhao et al. 2015; Fite and Gomez-Cambronero 2016). Studies by Wellner and colleagues showed that miR-203 is under control of ZEB1, which acts as both an inducer of the TGFβ-related EMT as well as a mediator of differentiation and self-renewal of CSC. Thus, by repressing stemness-inhibiting microRNAs, i.e. miR-200, miR-203 and miR-183, ZEB1 promotes tumorigenicity of the cells (Wellner et al. 2009).
MiR-205 is one of the critical regulator of stemness, also in breast cancer cells. Its physiological role is to supress ZEB1/2 expression, preventing EMT processes and maintaining differentiated state of cells. MiR-205 is repressed by the ligand Jagged1, a stroma-derived factor, promoting cancer stem cell-like phenotype (Lu et al. 2013; Chao et al. 2014). Silencing of miR-205 in mammary epithelial cells stimulated EMT, disrupted epithelial cell polarity and expanded stem cell population (Lu et al. 2013; Chao et al. 2014). Interestingly, in vivo studies indicated that miR-205-deficient mice spontaneously developed mammary lesions, while activation of miR-205 markedly diminished breast cancer cell stemness (Bojmar et al. 2013; Chao et al. 2014). In TNBC, downregulation of miR-205 resulted in chemoresistance, mainly due to induction of EMT and stemness (Sempere et al. 2007).
MiR-21 overexpression, identified in many solid tumours, is best characterised in TNBC (Table 1). Existing evidence demonstrates that TGFβ stimulation increases miR-21 expression in cancer cells, which in turn upregulates EMT process. This is associated with induction of BCSC-like phenotype and increase of hypoxia-inducible factor (HIF1α) levels. MiR-21 targets many different gene transcripts, such as PDCD4, PTEN, HIF1α, TIMP3 or TM1 mRNAs (Table 1) (Lu et al. 2008; Qi et al. 2009; Huang et al. 2009; Han et al. 2012; Mattos-Arruda et al. 2015). Han et al. observed that breast cancer stem cells undergone EMT express higher miR-21 levels than BCa cells not subjected to EMT. Interestingly, downregulation of miR-21 in BCSCs leads to MET, decrease of HIF1α and suppression of cell migration and invasion (Han et al. 2012). Although a direct association between miR-21 and EMT requires more thorough investigation, this microRNA seems to be of particular importance for BCa pathophysiology and may serve as a good indicator of treatment efficacy. Interestingly, in addition to its presumptive function in TNBC cells, high miR-21 level in tumour stroma was found to be also associated with poor disease outcome of the patients (MacKenzie et al. 2014).
Migration, invasion and metastasis
MiR-145 is representative of microRNAs that regulate cells migration, invasion, and metastasis (Table 1). In BCa cells, it targets mucin-1 (MUC1) and c-MYC—mRNA, both associated with cell invasiveness (Sachdeva and Mo 2010). MiR-145 downregulation is detected in approximately 10% of invasive breast carcinomas. MiR-145 was also shown to regulate invasion in TNBC by regulating ARF6 protein (Eades et al. 2015). Recent studies indicate that upregulation of miR-145 significantly reduces cell motility in MDA-MB-231 cells via targeting junctional adhesion molecule A (JAMA-A) and fascin as well as through effect on expression pattern of several motility-related proteins such as ROCK1, FSCN1 or TRMP3 involved in regulation of actin stress fibres or formation of filopodia (Sachdeva and Mo 2010; Götte et al. 2010).
MiR-373 is frequently upregulated in TNBC tissue and blood serum. In contrast to miR-145, its downregulation was found to impair cell migration and invasiveness (Table 1) (Huang et al. 2008; Eichelser et al. 2013; Chen et al. 2015). MiR-373 targets transcripts of CD44 and TXNIP, and activates two important EMT-inducers, HIF1α and Twist (mainly by targeting TXINP, identified as a metastasis suppressor), which in turn, in a positive feedback loop, upregulates miR-373 expression. Chen et al. showed that upregulation of miR-373-TXINP-HIF1α-Twist axis correlated with poor outcome of breast cancer patients. This suggests that activation of this signalling pathway may serve as both a potential biomarker and a new therapeutic target (Chen et al. 2015).
MiR-10b is highly expressed in TNBC cell lines (MDA-MB-231 and SUM1315), when compared to normal mammary epithelial (HMECS, MCF10A) or tumourigenic, but non-metastatic cells (SUM149 or SUM159), and enhances metastatic potential of cells grown in xenografts (Ma et al. 2007; Edmonds et al. 2009). MiR-10b positively regulates cell migration and invasion as well as influences expression of miR-9 (Table 1). High level of TGFβ was associated with upregulation of miR-10b in TNBC cell lines, whereas inhibition of miR-10b partially reversed EMT, and suppressed cell motility and proliferation (Han et al. 2014). MiR-10b was also reported to be positively correlated with Twist and was considered as an important mediator of twist-induced motility and invasiveness (Ma et al. 2007).
Prognostic value of miRNAs in TNBC
Despite many attempts in development of personalized therapy among molecular subtypes of TNBC, no breakthrough has been achieved yet. The guidelines for treatment of TNBC patients still encompass conventional surgery, radiotherapy, and chemotherapy (individually or in combination) (Costa and Gradishar 2017). Although some reports suggest that early response to specific chemotherapeutic regimens of TNBC is better than other BCa subtypes, TNBC patients are doomed to poor prognosis and chemoresistance (Pareja et al. 2016). The pattern of several microRNAs is substantially altered in TNBC suggesting they are likely to serve as useful prognostic factors in the disease (Dong et al. 2014; Sahlberg et al. 2015; Liu et al. 2017; Lü et al. 2017). For instance, decreased expression of miR-155 predicted poor overall survival in TNBC patients, while elevated levels of miR-21, miR-27a/b, miR-210, and miR-454 were associated with shorter overall survival (Medimegh et al. 2014; Sahlberg et al. 2015; Thakur et al. 2016; Lü et al. 2017). Similarly, decreased expression of miR-374a/b and increased level of miR-454 correlated with shorter disease-free survival (Radojicic et al. 2011).
Other panels of miRNAs was identified to be associated with chemoresistance (Gasparini et al. 2014; Ouyang et al. 2014; Shen et al. 2014; Sahlberg et al. 2015). For example, expression of miR-181a was elevated in TNBC tissue samples from patients who did not respond to neo-adjuvant chemotherapy and was significantly inversely correlated with chemo-sensitivity (Ouyang et al. 2014). In vitro studies by Ouyang et al. demonstrated that in the MDA-MB-231 cell line as well as in TNBC tissue samples, upregulation of miR-155-5p, miR-21-3p, miR-181a-5p, miR-181b-5p, miR-183-5p and downregulation of miR-10b-5p, miR-451a, miR-125b-5p, miR-31-5p, miR-195-5p were associated with chemoresistance to doxorubicin (Korpal et al. 2008; Chen et al. 2012; Kong et al. 2014; Liu et al. 2015; Fkih et al. 2017). Low level of miR-200c was shown to be connected with resistance to doxorubicin, poor response to radiotherapy and elevated expression of multidrug resistance gene (Korpal et al. 2008). Identification of miRNA clusters, whose deregulated levels accompany resistance to chemotherapy, may open new avenues in development of more efficient therapies.
Conclusions
This review focused on several microRNAs shown to be specific to triple-negative breast carcinoma. Their role in TNBC biology was discussed in relation to molecular processes underlying disease progression, with particular emphasis on the epithelial-to-mesenchymal transition. Although, for the most part, the degree of involvement in TNBC pathophysiology remains to be established, increasing evidence suggests that specific microRNA clusters might be of clinical utility as both predictive markers and potential therapeutic targets in this highly aggressive form of breast cancer.
References
Arpino G, Milano M, De Placido S (2015) Features of aggressive breast cancer. Breast 24:594–600. https://doi.org/10.1016/J.BREAST.2015.06.001
Avery-Kiejda KA, Mathe A, Scott RJ (2017) Genome-wide miRNA, gene and methylation analysis of triple negative breast cancer to identify changes associated with lymph node metastases. Genom Data 14:1–4. https://doi.org/10.1016/J.GDATA.2017.07.004
Baroni S, Romero-Cordoba S, Plantamura I et al (2016) Exosome-mediated delivery of miR-9 induces cancer-associated fibroblast-like properties in human breast fibroblasts. Cell Death Dis 7:e2312–e2312. https://doi.org/10.1038/cddis.2016.224
Berber U, Yilmaz I, Narli G, Haholu A, Kucukodaci Z, Demirel D (2014) miR-205 and miR-200c: predictive micro RNAs for lymph node metastasis in triple negative breast cancer. J Breast Cancer 17(2):143–148. https://doi.org/10.4048/jbc.2014.17.2.143
Bojmar L, Karlsson E, Ellegård S et al (2013) The role of MicroRNA-200 in progression of human colorectal and breast cancer. PLoS One 8:e84815. https://doi.org/10.1371/journal.pone.0084815
Chang YY, Kuo WH, Hung JH et al (2015) Deregulated microRNAs in triple-negative breast cancer revealed by deep sequencing. Mol Cancer 14:36. https://doi.org/10.1186/s12943-015-0301-9
Chao C-H, Chang C-C, Wu M-J et al (2014) MicroRNA-205 signaling regulates mammary stem cell fate and tumorigenesis. J Clin Invest 124:3093–3106. https://doi.org/10.1172/JCI73351
Chen J, Tian W, Cai H et al (2012) Down-regulation of microRNA-200c is associated with drug resistance in human breast cancer. Med Oncol 29:2527–2534. https://doi.org/10.1007/s12032-011-0117-4
Chen D, Dang B-L, Huang J et al (2015) MiR-373 drives the epithelial-to-mesenchymal transition and metastasis via the miR-373-TXNIP-HIF1α-TWIST signaling axis in breast cancer. Oncotarget 6:32701–32712. https://doi.org/10.18632/oncotarget.4702
Chen J, Shin VY, Siu MT et al (2016) miR-199a-5p confers tumor-suppressive role in triple-negative breast cancer. BMC Cancer 16:887. https://doi.org/10.1186/s12885-016-2916-7
Cohen EN, Gao H, Anfossi S et al (2015) Inflammation mediated metastasis: immune induced epithelial-to-mesenchymal transition in inflammatory breast cancer cells. PLoS One 10:1–18. https://doi.org/10.1371/journal.pone.0132710
Costa RLB, Gradishar WJ (2017) Triple-negative breast cancer: current practice and future directions. J Oncol Pract 13:301–303. https://doi.org/10.1200/JOP.2017.023333
D’Angelo RC, Ouzounova M, Davis A et al (2015) Notch reporter activity in breast cancer cell lines identifies a subset of cells with stem cell activity. Mol Cancer Ther 14:779–787. https://doi.org/10.1158/1535-7163.MCT-14-0228
D’Ippolito E, Plantamura I, Bongiovanni L et al (2016) miR-9 and miR-200 regulate PDGFRβ-mediated endothelial differentiation of tumor cells in triple-negative breast cancer. Cancer Res 76:5562–5572. https://doi.org/10.1158/0008-5472.CAN-16-0140
DeCastro AJ, Dunphy KA, Hutchinson J et al (2013) MiR203 mediates subversion of stem cell properties during mammary epithelial differentiation via repression of ∆NP63α and promotes mesenchymal-to-epithelial transition. Cell Death Dis 4:e514. https://doi.org/10.1038/cddis.2013.37
Dong G, Liang X, Wang D et al (2014) High expression of miR-21 in triple-negative breast cancers was correlated with a poor prognosis and promoted tumor cell in vitro proliferation. Med Oncol 31:57. https://doi.org/10.1007/s12032-014-0057-x
Drago-Ferrante R, Pentimalli F, Carlisi D, De Blasio A, Saliba C, Baldacchino S, Degaetano J, Debono J, Caruana-Dingli G, Grech G, Scerri C, Tesoriere G, Giordano A, Vento R, Di Fiore R (2017) Suppressive role exerted by microRNA-29b-1-5p in triple negative breast cancer through SPIN1 regulation. Oncotarget 8(17):28939–28958. https://doi.org/10.18632/oncotarget.15960
Eades G, Wolfson B, Zhang Y et al (2015) lincRNA-RoR and miR-145 regulate invasion in triple-negative breast cancer via targeting ARF6. Mol Cancer Res 13:330–338. https://doi.org/10.1158/1541-7786.MCR-14-0251
Edmonds MD, Hurst DR, Vaidya KS et al (2009) Breast cancer metastasis suppressor 1 coordinately regulates metastasis-associated microRNA expression. Int J cancer 125:1778–1785. https://doi.org/10.1002/ijc.24616
Eichelser C, Flesch-Janys D, Chang-Claude J et al (2013) Deregulated serum concentrations of circulating cell-free microRNAs miR-17, miR-34a, miR-155, and miR-373 in human breast cancer development and progression. Clin Chem 59:1489–1496. https://doi.org/10.1373/clinchem.2013.205161
Erturk E, Cecener G, Tezcan G et al (2015) BRCA mutations cause reduction in miR-200c expression in triple negative breast cancer. Gene 556:163–169. https://doi.org/10.1016/j.gene.2014.11.047
Espinoza I, Pochampally R, Xing F et al (2013) Notch signaling: targeting cancer stem cells and epithelial-to-mesenchymal transition. Onco Targets Ther 6:1249–1259. https://doi.org/10.2147/OTT.S36162
Felipe Lima J, Nofech-Mozes S, Bayani J, Bartlett JMS (2016) EMT in breast carcinoma—a review. J Clin Med 5:65. https://doi.org/10.3390/jcm5070065
Ferlay J, Soerjomataram II, Dikshit R et al (2014) Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer 136:E359-86. https://doi.org/10.1002/ijc.29210
Fire A, Xu S, Montgomery MK et al (1998) Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391:806–811. https://doi.org/10.1038/35888
Fite K, Gomez-Cambronero J (2016) Down-regulation of MicroRNAs (MiRs) 203, 887, 3619 and 182 prevents vimentin-triggered, phospholipase D (PLD)-mediated cancer cell invasion. J Biol Chem 291:719–730. https://doi.org/10.1074/jbc.M115.686006
Fkih M, Privat I, Trimeche M M, et al (2017) miR-10b, miR-26a, miR-146a And miR-153 expression in triple negative vs non triple negative breast cancer: potential biomarkers. Pathol Oncol Res 23:815–827. https://doi.org/10.1007/s12253-017-0188-4
Gasparini P, Cascione L, Fassan M et al (2014) microRNA expression profiling identifies a four microRNA signature as a novel diagnostic and prognostic biomarker in triple negative breast cancers. Oncotarget 5:1174–1184. https://doi.org/10.18632/oncotarget.1682
Gomes BC, Martins M, Lopes P, Morujão I, Oliveira M, Araújo A, Rueff J, Rodrigues AS (2016) Prognostic value of microRNA-203a expression in breast cancer. Oncol Rep 36(3):1748–1756. https://doi.org/10.3892/or.2016.4913
Götte M, Mohr C, Koo C-Y et al (2010) miR-145-dependent targeting of Junctional Adhesion Molecule A and modulation of fascin expression are associated with reduced breast cancer cell motility and invasiveness. Oncogene 29:6569–6580. https://doi.org/10.1038/onc.2010.386
Gregory PA, Bert AG, Paterson EL et al (2008) The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol 10:593–601. https://doi.org/10.1038/ncb1722
Han M, Wang Y, Liu M et al (2012) MiR-21 regulates epithelial-mesenchymal transition phenotype and hypoxia-inducible factor-1α expression in third-sphere forming breast cancer stem cell-like cells. Cancer Sci 103:1058–1064. https://doi.org/10.1111/j.1349-7006.2012.02281.x
Han X, Yan S, Weijie Z et al (2014) Critical role of miR-10b in transforming growth factor-β1-induced epithelial–mesenchymal transition in breast cancer. Cancer Gene Ther 21:60–67. https://doi.org/10.1038/cgt.2013.82
Hannon GJ, Hammond SM, Bernstein E, Beach D (2000) An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature 404:293–296. https://doi.org/10.1038/35005107
Hill L, Browne G, Tulchinsky E (2013) ZEB/miR-200 feedback loop: at the crossroads of signal transduction in cancer. Int J Cancer 132(4):745–754. https://doi.org/10.1002/ijc.27708
Hollier BG, Tinnirello AA, Werden SJ et al (2013) FOXC2 expression links epithelial-mesenchymal transition and stem cell properties in breast cancer. Cancer Res 73:1981–1992. https://doi.org/10.1158/0008-5472.CAN-12-2962
Huang Q, Gumireddy K, Schrier M et al (2008) The microRNAs miR-373 and miR-520c promote tumour invasion and metastasis. Nat Cell Biol 10:202–210. https://doi.org/10.1038/ncb1681
Huang T-H, Wu F, Loeb GB et al (2009) Up-regulation of miR-21 by HER2/neu signaling promotes cell invasion. J Biol Chem 284:18515–18524. https://doi.org/10.1074/jbc.M109.006676
Humphries B, Wang Z, Oom AL et al (2014) MicroRNA-200b targets protein kinase Cα and suppresses triple-negative breast cancer metastasis. Carcinogenesis 35:2254–2263. https://doi.org/10.1093/carcin/bgu133
Huo L, Wang Y, Gong Y et al (2016) MicroRNA expression profiling identifies decreased expression of miR-205 in inflammatory breast cancer. Mod Pathol 29:330–346. https://doi.org/10.1038/modpathol.2016.38
Hutvágner G, McLachlan J, Pasquinelli AE et al (2001) A cellular function for the RNA-interference enzyme dicer in the maturation of the let-7 small temporal RNA. Science 293:834–838. https://doi.org/10.1126/science.1062961
Hwang H-W, Mendell JT (2006) MicroRNAs in cell proliferation, cell death, and tumorigenesis. Br J Cancer 94:776–780. https://doi.org/10.1038/sj.bjc.6603023
Hwang MS, Yu N, Stinson SY et al (2013) miR-221/222 targets adiponectin receptor 1 to promote the epithelial-to-mesenchymal transition in breast cancer. PLoS One 8:e66502. https://doi.org/10.1371/journal.pone.0066502
Jang G-B, Kim J-Y, Cho S-D et al (2015) Blockade of Wnt/β-catenin signaling suppresses breast cancer metastasis by inhibiting CSC-like phenotype. Sci Rep 5:12465. https://doi.org/10.1038/srep12465
Jang MH, Kim HJ, Gwak JM et al (2017) Prognostic value of microRNA-9 and microRNA-155 expression in triple-negative breast cancer. Hum Pathol 68:69–78. https://doi.org/10.1016/j.humpath.2017.08.026
Jurmeister S, Baumann M, Balwierz A et al (2012) MicroRNA-200c represses migration and invasion of breast cancer cells by targeting actin-regulatory proteins FHOD1 and PPM1F. Mol Cell Biol 32:633–651. https://doi.org/10.1128/MCB.06212-11
Kaidi A, Williams AC, Paraskeva C (2007) Interaction between β-catenin and HIF-1 promotes cellular adaptation to hypoxia. Nat Cell Biol 9:210–217. https://doi.org/10.1038/ncb1534
Kalluri R, Weinberg RA (2009) The basics of epithelial-mesenchymal transition. J Clin Invest 119:1420–1428. https://doi.org/10.1172/JCI39104
Kolacinska A, Morawiec J, Fendler W et al (2014) Association of microRNAs and pathologic response to preoperative chemotherapy in triple negative breast cancer: preliminary report. Mol Biol Rep 41:2851–2857. https://doi.org/10.1007/s11033-014-3140-7
Kong W, He L, Richards EJ et al (2014) Upregulation of miRNA-155 promotes tumour angiogenesis by targeting VHL and is associated with poor prognosis and triple-negative breast cancer. Oncogene 33:679–689. https://doi.org/10.1038/onc.2012.636
Korpal M, Lee ES, Hu G, Kang Y (2008) The miR-200 family inhibits epithelial-mesenchymal transition and cancer cell migration by direct targeting of E-cadherin transcriptional repressors ZEB1 and ZEB2. J Biol Chem 283:14910–14914. https://doi.org/10.1074/jbc.C800074200
Kurozumi S, Yamaguchi Y, Kurosumi M, Ohira M, Matsumoto H, Horiguchi J (2017) Recent trends in microRNA research into breast cancer with particular focus on the associations between microRNAs and intrinsic subtypes. J Human Genet 62:15–24. https://doi.org/10.1038/jhg.2016.89
Lamouille S, Subramanyam D, Blelloch R, Derynck R (2013) Regulation of epithelial-mesenchymal and mesenchymal-epithelial transitions by microRNAs. Curr Opin Cell Biol 25:200–207. https://doi.org/10.1016/j.ceb.2013.01.008
Lamouille S, Xu J, Derynck R (2014) Molecular mechanisms of epithelial-mesenchymal transition. Nat Rev Mol Cell Biol 15:178–196. https://doi.org/10.1038/nrm3758
Lee Y, Jeon K, Lee JT et al (2002) MicroRNA maturation: stepwise processing and subcellular localization. EMBO J 21:4663–4670. https://doi.org/10.1093/emboj/cdf476
Lee Y, Ahn C, Han J et al (2003) The nuclear RNase III Drosha initiates microRNA processing. Nature 425:415–419. https://doi.org/10.1038/nature01957
Lehmann BD, Pietenpol JA (2015) Clinical implications of molecular heterogeneity in triple negative breast cancer. Breast 24(Suppl 2):S36–S40. https://doi.org/10.1016/j.breast.2015.07.009
Leibovich-Rivkin T, Liubomirski Y, Bernstein B et al (2013) Inflammatory factors of the tumor microenvironment induce plasticity in nontransformed breast epithelial cells: EMT, invasion, and collapse of normally organized breast textures. Neoplasia 15:1330–1346. https://doi.org/10.1593/neo.131688
Liang Y, Yang W, Zhu Y, Yuan Y (2016) Prognostic role of microRNA-203 in various carcinomas: evidence from a meta-analysis involving 13 studies. Springerplus 5(1):1538. https://doi.org/10.1186/s40064-016-3225-y
Lim S, Becker A, Zimmer A et al (2013) SNAI1-mediated epithelial-mesenchymal transition confers chemoresistance and cellular plasticity by regulating genes involved in cell death and stem cell maintenance. PLoS One 8:e66558. https://doi.org/10.1371/journal.pone.0066558
Lin S, Gregory RI (2015) MicroRNA biogenesis pathways in cancer. Nat Rev Cancer 15:321–333. https://doi.org/10.1038/nrc3932
Liu P, Tang H, Chen B et al (2015) MiR-26a suppresses tumour proliferation and metastasis by targeting metadherin in triple negative breast cancer. Cancer Lett 357:384–392. https://doi.org/10.1016/j.canlet.2014.11.050
Liu Y, Zhang Y, Li Q et al (2017) MiRNAs predict the prognosis of patients with triple negative breast cancer: a meta-analysis. PLoS One 12:e0170088. https://doi.org/10.1371/journal.pone.0170088
López-Novoa JM, Nieto, MA (2009) Inflammation and EMT: an alliance towards organ fibrosis and cancer progression Introduction—EMT in cancer and fibrosis. EMBO Mol Med 303–314. https://doi.org/10.1002/emmm.200900043
Lu Z, Liu M, Stribinskis V et al (2008) MicroRNA-21 promotes cell transformation by targeting the programmed cell death 4 gene. Oncogene 27:4373–4379. https://doi.org/10.1038/onc.2008.72
Lu J, Ye X, Fan F et al (2013) Endothelial cells promote the colorectal cancer stem cell phenotype through a soluble form of jagged-1. Cancer Cell 23:171–185. https://doi.org/10.1016/j.ccr.2012.12.021
Lü L, Mao X, Shi P et al (2017) MicroRNAs in the prognosis of triple-negative breast cancer. Med (United States) 96:e7085. https://doi.org/10.1097/MD.0000000000007085
Ma L, Teruya-Feldstein J, Weinberg RA (2007) Tumour invasion and metastasis initiated by microRNA-10b in breast cancer. Nature 449:682–688. https://doi.org/10.1038/nature06174
Ma L, Young J, Prabhala H et al (2010) miR-9, a MYC/MYCN-activated microRNA, regulates E-cadherin and cancer metastasis. Nat Cell Biol 12:247–256. https://doi.org/10.1038/ncb2024
MacKenzie TA, Schwartz GN, Calderone HM et al (2014) Stromal expression of miR-21 identifies high-risk group in triple-negative breast cancer. Am J Pathol 184:3217–3225. https://doi.org/10.1016/J.AJPATH.2014.08.020
Madhavan D, Cuk K, Burwinkel B, Yang R (2013) Cancer diagnosis and prognosis decoded by blood-based circulating microRNA signatures. Front Genet 4:116. https://doi.org/10.3389/fgene.2013.00116
Mantovani A, Allavena P, Sica A, Balkwill F (2008) Cancer-related inflammation. Nature 454:436–444. https://doi.org/10.1038/nature07205
Martinez J, Patkaniowska A, Urlaub H et al (2002) Single-stranded antisense siRNAs guide target RNA cleavage in RNAi. Cell 110:563–574. https://doi.org/10.1016/S0092-8674(02)00908-X
Massagué J (2012) TGFβ signalling in context. Nat Rev Mol Cell Biol 13:616–630. https://doi.org/10.1038/nrm3434
Mattos-Arruda L, De Bottai G, Nuciforo PG et al (2015) MicroRNA-21 links epithelial-to-mesenchymal transition and inflammatory signals to confer resistance to neoadjuvant trastuzumab and chemotherapy in HER2-positive breast cancer patients. Oncotarget 6:37269–37280. https://doi.org/10.18632/oncotarget.5495
Medimegh I, Omrane I, Privat M et al (2014) MicroRNAs expression in triple negative vs non triple negative breast cancer in Tunisia: interaction with clinical outcome. PLoS One 9:e111877. https://doi.org/10.1371/journal.pone.0111877
Mekala JR, Naushad SM, Ponnusamy L et al (2018) Epigenetic regulation of miR-200 as the potential strategy for the therapy against triple-negative breast cancer. Gene 641:248–258. https://doi.org/10.1016/J.GENE.2017.10.018
Nassirpour R, Mehta PP, Baxi SM, Yin M-J (2013) miR-221 promotes tumorigenesis in human triple negative breast cancer cells. PLoS One 8:e62170. https://doi.org/10.1371/journal.pone.0062170
Niehrs C (2012) The complex world of WNT receptor signalling. Nat Rev Mol Cell Biol 13:767–779. https://doi.org/10.1038/nrm3470
Olivieri F, Rippo MR, Monsurrò V et al (2013) MicroRNAs linking inflamm-aging, cellular senescence and cancer. Ageing Res Rev 12:1056–1068. https://doi.org/10.1016/j.arr.2013.05.001
Ouyang M, Li Y, Ye S et al (2014) MicroRNA profiling implies new markers of chemoresistance of triple-negative breast cancer. PLoS One 9:e96228. https://doi.org/10.1371/journal.pone.0096228
Pareja F, Geyer FC, Marchiò C et al (2016) Triple-negative breast cancer: the importance of molecular and histologic subtyping, and recognition of low-grade variants. Breast Cancer 2:16036. https://doi.org/10.1038/npjbcancer.2016.36
Paszek S, Gabło N, Barnaś E et al (2017) Dysregulation of microRNAs in triple-negative breast cancer. Ginekol Pol 88:530–536. https://doi.org/10.5603/GP.a2017.0097
Pecot CV, Rupaimoole R, Yang D, Akbani R, Ivan C, Lu C, Wu S, Han H-D, Shah MY, Rodriguez-Aguayo C, Bottsford-Miller J, Liu Y, Kim SB, Unruh A, Gonzalez-Villasana V, Huang L, Zand B, Moreno-Smith M, Mangala LS, Taylor M, Dalton HJ, Sehgal V, Wen Y, Kang Y, Baggerly KA, Lee J-S, Ram PT, Ravoori MK, Kundra V, Zhang X, Ali-Fehmi R, Gonzalez-Angulo A-M, Massion PP, Calin GA, Lopez-Berestein G, Zhang W, Sood AK (2013) Tumour angiogenesis regulation by the miR-200 family. Nat Commun 4:2427. https://doi.org/10.1038/ncomms3427
Perou CM, Sørlie T, Eisen MB et al (2000) Molecular portraits of human breast tumours. Nature 406:747–752. https://doi.org/10.1038/35021093
Piovan C, Palmieri D, Di Leva G et al (2012) Oncosuppressive role of p53-induced miR-205 in triple negative breast cancer. Mol Oncol 6:458–472. https://doi.org/10.1016/j.molonc.2012.03.003
Polyak K (2007) Science in medicine Breast cancer: origins and evolution. Cell 117:3155–3163. https://doi.org/10.1172/JCI33295.group
Qi L, Bart J, Tan LP et al (2009) Expression of miR-21 and its targets (PTEN, PDCD4, TM1) in flat epithelial atypia of the breast in relation to ductal carcinoma in situ and invasive carcinoma. BMC Cancer 9:163. https://doi.org/10.1186/1471-2407-9-163
Radojicic J, Zaravinos A, Vrekoussis T et al (2011) MicroRNA expression analysis in triple-negative (ER, PR and Her2/neu) breast cancer. Cell Cycle 10:507–517. https://doi.org/10.4161/cc.10.3.14754
Rhodes LV, Martin EC, Segar HC et al (2015) Dual regulation by microRNA-200b-3p and microRNA-200b-5p in the inhibition of epithelial-to-mesenchymal transition in triple-negative breast cancer. Oncotarget 6:16638–16652. https://doi.org/10.18632/oncotarget.3184
Ricciardi M, Zanotto M, Malpeli G et al (2015) Epithelial-to-mesenchymal transition (EMT) induced by inflammatory priming elicits mesenchymal stromal cell-like immune-modulatory properties in cancer cells. Br J Cancer 112:1067–1075. https://doi.org/10.1038/bjc.2015.29
Sachdeva M, Mo Y-Y (2010) MicroRNA-145 suppresses cell invasion and metastasis by directly targeting mucin 1. Cancer Res 70:378–387. https://doi.org/10.1158/0008-5472.CAN-09-2021
Sahlberg KK, Bottai G, Naume B et al (2015) A serum MicroRNA signature predicts tumor relapse and survival in triple-negative breast cancer patients. Clin Cancer Res 21:1207–1214. https://doi.org/10.1158/1078-0432.CCR-14-2011
Sahlgren C, Gustafsson MV, Jin S et al (2008) Notch signaling mediates hypoxia-induced tumor cell migration and invasion. Proc Natl Acad Sci 105:6392–6397. https://doi.org/10.1073/pnas.0802047105
Scheel C, Weinberg RA (2012) Cancer stem cells and epithelial-mesenchymal transition: concepts and molecular links. Semin Cancer Biol 22:396–403. https://doi.org/10.1016/j.semcancer.2012.04.001
Sempere LF, Christensen M, Silahtaroglu A et al (2007) Altered MicroRNA expression confined to specific epithelial cell subpopulations in breast cancer. Cancer Res 67:11612–11620. https://doi.org/10.1158/0008-5472.CAN-07-5019
Sethi S, Sarkar FH, Ahmed Q et al (2011) Molecular markers of epithelial-to-mesenchymal transition are associated with tumor aggressiveness in breast carcinoma. Transl Oncol 4:222–226. https://doi.org/10.1593/tlo.10244
Seton-Rogers S (2016) Epithelial-mesenchymal transition: untangling EMT’s functions. Nat Rev Cancer 16:1. https://doi.org/10.1038/nrc.2015.6
Shen S, Sun Q, Liang Z et al (2014) A prognostic model of triple-negative breast cancer based on miR-27b-3p and node status. PLoS One 9:e100664. https://doi.org/10.1371/journal.pone.0100664
Shirakihara T, Horiguchi K, Miyazawa K et al (2011) TGF-β regulates isoform switching of FGF receptors and epithelial-mesenchymal transition. EMBO J 30:783–795. https://doi.org/10.1038/emboj.2010.351
Shivdasani RA (2006) MicroRNAs: regulators of gene expression and cell differentiation. Blood 108:3646–3653. https://doi.org/10.1182/blood-2006-01-030015
Shostak K, Chariot A (2011) NF-κB, stem cells and breast cancer: the links get stronger. Breast Cancer Res 13:214. https://doi.org/10.1186/bcr2886
Singh A, Settleman J (2010) EMT, cancer stem cells and drug resistance: an emerging axis of evil in the war on cancer. Oncogene 29:4741–4751. https://doi.org/10.1038/onc.2010.215
Stinson S, Lackner MR, Adai AT et al (2011) miR-221/222 targeting of trichorh inophalangeal 1 (TRPS1) promotes epithelial-to-mesenchymal transition in breast cancer. Sci Signal 4(186):pt5. https://doi.org/10.1126/scisignal.2002258
Tao Z, Shi A, Lu C et al (2015) Breast cancer: epidemiology and etiology. Cell Biochem Biophys 72:333–338. https://doi.org/10.1007/s12013-014-0459-6
Thakur S, Grover RK, Gupta S et al (2016) Identification of specific miRNA signature in paired sera and tissue samples of Indian women with triple negative breast cancer. PLoS One 11:e0158946. https://doi.org/10.1371/journal.pone.0158946
Thiery JP, Acloque H, Huang RYJ, Nieto MA (2009) Epithelial-mesenchymal transitions in development and disease. Cell 139:871–890. https://doi.org/10.1016/j.cell.2009.11.007
Wang Y, Zhou BP (2011) Epithelial-mesenchymal transition in breast cancer progression and metastasis. Chin J Cancer 30:603–611. https://doi.org/10.5732/cjc.011.10226
Wang J, Zhao H, Tang D et al (2013) Overexpressions of microRNA-9 and microRNA-200c in human breast cancers are associated with lymph node metastasis. Cancer Biother Radiopharm 28:283–288. https://doi.org/10.1089/cbr.2012.1293
Wang J, Tsouko E, Jonsson P et al (2014) miR-206 inhibits cell migration through direct targeting of the actin-binding protein Coronin 1C in triple-negative breast cancer. Mol Oncol 8:1690–1702. https://doi.org/10.1016/j.molonc.2014.07.006
Wang S-S, Jiang J, Liang X-H, Tang Y-L (2015) Links between cancer stem cells and epithelial-mesenchymal transition. Onco Targets Ther 8:2973–2980. https://doi.org/10.2147/OTT.S91863
Weiss A, Attisano L (2013) The TGFbeta superfamily signaling pathway. Wiley Interdiscip Rev Dev Biol 2:47–63. https://doi.org/10.1002/wdev.86
Wellner U, Schubert J, Burk UC et al (2009) The EMT-activator ZEB1 promotes tumorigenicity by repressing stemness-inhibiting microRNAs. Nat Cell Biol 11:1487–1495. https://doi.org/10.1038/ncb1998
Wu Y, Deng J, Rychahou PG et al (2009) Stabilization of snail by NF-κB is required for inflammation-induced cell migration and invasion. Cancer Cell 15:416–428. https://doi.org/10.1016/j.ccr.2009.03.016
Yamamoto M, Taguchi Y, Ito-Kureha T et al (2013) NF-κB non-cell-autonomously regulates cancer stem cell populations in the basal-like breast cancer subtype. Nat Commun 4:2299. https://doi.org/10.1038/ncomms3299
Yang J, Zhang J, Chen J, Xu Y, Song N-H, Yin C-J, Zhang L (2014) Prognostic role of microRNA-221 in various human malignant neoplasms: a meta-analysis of 20 related studies. PLoS One 9(1):e87606. https://doi.org/10.1371/journal.pone.0087606
Yook JI, Li X-Y, Ota I et al (2006) A Wnt–Axin2–GSK3β cascade regulates Snail1 activity in breast cancer cells. Nat Cell Biol 8:1398–1406. https://doi.org/10.1038/ncb1508
Yuan T, Wu Y, Zhou BP (2012) Tumor inflammatory microenvironment in EMT and metastasis. Tumor Microenviron Myelomonocyt Cells. https://doi.org/10.5772/32219
Zaravinos A, Apostolos (2015) The regulatory role of MicroRNAs in EMT and cancer. J Oncol 2015:865816. https://doi.org/10.1155/2015/865816
Zhao S, Han J, Zheng L et al (2015) MicroRNA-203 regulates growth and metastasis of breast cancer. Cell Physiol Biochem 37:35–42. https://doi.org/10.1159/000430331
Zhu H, Dai M, Chen X et al (2017) Integrated analysis of the potential roles of miRNA-mRNA networks in triple negative breast cancer. Mol Med Rep 16:1139–1146. https://doi.org/10.3892/mmr.2017.6750
Acknowledgements
This work was supported by the Polish National Science Centre: UMO-2015/17/B/ NZ4/02157 and by Medical University of Lodz: 502-03/1-034-03/502-14-329, 502-03/1-034-03/502-14-330.
Funding
This study was funded by the following grants: Polish National Science Centre: UMO-2015/17/B/ NZ4/02157; Medical University of Lodz: 502-03/1-034-03/502-14-329 and 502-03/1-034-03/502-14-330.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Piasecka, D., Braun, M., Kordek, R. et al. MicroRNAs in regulation of triple-negative breast cancer progression. J Cancer Res Clin Oncol 144, 1401–1411 (2018). https://doi.org/10.1007/s00432-018-2689-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00432-018-2689-2