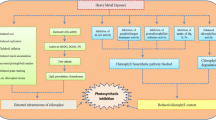
Abstract
Main conclusion
Arundo donax ecotypes react differently to salinity, partly due to differences in constitutive defences and methylome plasticity.
Abstract
Arundo donax L. is a C3 fast-growing grass that yields high biomass under stress. To elucidate its ability to produce biomass under high salinity, we investigated short/long-term NaCl responses of three ecotypes through transcriptional, metabolic and DNA methylation profiling of leaves and roots. Prolonged salt treatment discriminated the sensitive ecotype ‘Cercola’ from the tolerant ‘Domitiana’ and ‘Canneto’ in terms of biomass. Transcriptional and metabolic responses to NaCl differed between the ecotypes. In roots, constitutive expression of ion transporter and stress-related transcription factors’ genes was higher in ‘Canneto’ and ‘Domitiana’ than ‘Cercola’ and 21-day NaCl drove strong up-regulation in all ecotypes. In leaves, unstressed ‘Domitiana’ confirmed higher expression of the above genes, whose transcription was repressed in ‘Domitiana’ but induced in ‘Cercola’ following NaCl treatment. In all ecotypes, salinity increased proline, ABA and leaf antioxidants, paralleled by up-regulation of antioxidant genes in ‘Canneto’ and ‘Cercola’ but not in ‘Domitiana’, which tolerated a higher level of oxidative damage. Changes in DNA methylation patterns highlighted a marked capacity of the tolerant ‘Domitiana’ ecotype to adjust DNA methylation to salt stress. The reduced salt sensitivity of ‘Domitiana’ and, to a lesser extent, ‘Canneto’ appears to rely on a complex set of constitutively activated defences, possibly due to the environmental conditions of the site of origin, and on higher plasticity of the methylome. Our findings provide insights into the mechanisms of adaptability of A. donax ecotypes to salinity, offering new perspectives for the improvement of this species for cultivation in limiting environments.





Similar content being viewed by others
Abbreviations
- MDA:
-
Malondialdehyde
- MSAP:
-
Methylation-sensitive amplification polymorphism
- TF:
-
Transcription factor
References
Aceituno FF, Moseyko N, Rhee SY, Gutiérrez RA (2008) The rules of gene expression in plants: organ identity and gene body methylation are key factors for regulation of gene expression in Arabidopsis thaliana. BMC Genom 9:438. https://doi.org/10.1186/1471-2164-9-438
Ahmad R, Liow PS, Spencer DF, Jasieniuk M (2008) Molecular evidence for a single genetic clone of invasive Arundo donax in the United States. Aquat Bot 88:113–120. https://doi.org/10.1016/j.aquabot.2007.08.015
Ahrar M, Doneva D, Koleva D et al (2015) Isoprene emission in the monocot Arundineae tribe in relation to functional and structural organization of the photosynthetic apparatus. Environ Exp Bot 119:87–95. https://doi.org/10.1016/j.envexpbot.2015.04.010
Aina R, Sgorbati S, Santagostino A et al (2004) Specific hypomethylation of DNA is induced by heavy metals in white clover and industrial hemp. Physiol Plant 121:472–480. https://doi.org/10.1111/j.1399-3054.2004.00343.x
Albertini E, Marconi G (2014) Methylation-sensitive amplified polymorphism (MSAP) marker to investigate drought-stress response in Montepulciano and Sangiovese grape cultivars. Methods Mol Biol 1112:151–164
Allu AD, Soja AM, Wu A et al (2014) Salt stress and senescence: identification of cross-talk regulatory components. J Exp Bot 65:3993–4008. https://doi.org/10.1093/jxb/eru173
Almeida DM, Margarida Oliveira M, Saibo NJMM et al (2017) Regulation of Na+ and K+ homeostasis in plants: towards improved salt stress tolerance in crop plants. Genet Mol Biol 40:326–345. https://doi.org/10.1590/1678-4685-GMB-2016-0106
Aversano R, Caruso I, Aronne G et al (2013) Stochastic changes affect Solanum wild species following autopolyploidization. J Exp Bot 64:625–635. https://doi.org/10.1093/jxb/ers357
Barrero RA, Guerrero FD, Moolhuijzen P, Goolsby JA, Tidwell J, Bellgard SE, Bellgard MI (2015) Shoot transcriptome of the giant reed, Arundo donax. Data Brief 3:1–6
Bernstein N, Kafkafi U (2002) Root growth under salinity stress. In: Waisel Y, Eshel A, Kafkafi U (eds) Plant roots: the hidden half, 3rd edn. Marcel Dekker, New York, pp 1194–1221
Borkotoky S, Saravanan V, Jaiswal A et al (2013) The Arabidopsis stress responsive gene database. Int J Plant Genom 2013:949564. https://doi.org/10.1155/2013/949564
Butelli E, Titta L, Giorgio M et al (2008) Enrichment of tomato fruit with health-promoting anthocyanins by expression of select transcription factors. Nat Biotechnol 26:1301–1308. https://doi.org/10.1038/nbt.1506
Choi CS, Sano H (2007) Abiotic-stress induces demethylation and transcriptional activation of a gene encoding a glycerophosphodiesterase-like protein in tobacco plants. Mol Genet Genomics 277:589. https://doi.org/10.1007/s00438-007-0209-1
Claussen W (2005) Proline as a measure of stress in tomato plants. Plant Sci 168:241–248. https://doi.org/10.1016/J.PLANTSCI.2004.07.039
Davenport RJ, Muñoz-Mayor A, Jha D et al (2007) The Na+ transporter AtHKT1;1 controls retrieval of Na+ from the xylem in Arabidopsis. Plant Cell Environ 30:497–507. https://doi.org/10.1111/j.1365-3040.2007.01637.x
De Palma M, Salzano M, Villano C et al (2019) Transcriptome reprogramming, epigenetic modifications and alternative splicing orchestrate the tomato root response to the beneficial fungus Trichoderma harzianum. Hort Res 6:5. https://doi.org/10.1038/s41438-018-0079-1
De Stefano R, Cappetta E, Guida G et al (2017) Screening of giant reed (Arundo donax L.) ecotypes for biomass production under salt stress. Plant Biosystems 152:911–917. https://doi.org/10.1080/11263504.2017.1362059
Deinlein U, Stephan AB, Horie T et al (2014) Plant salt-tolerance mechanisms. Trends Plant Sci 19:371–379. https://doi.org/10.1016/j.tplants.2014.02.001
DeRose-Wilson L, Gaut BS (2011) Mapping salinity tolerance during Arabidopsis thaliana germination and seedling growth. PLoS One 6(8):e22832. https://doi.org/10.1371/journal.pone.0022832
Dhindsa RS, Plumb-Dhindsa P, Thorpe TA (1981) Leaf senescence: correlated with increased levels of membrane permeability and lipid peroxidation, and decreased levels of superoxide dismutase and catalase. J Exp Bot 32(1):93–101
Di Mola I, Guida G, Mistretta C et al (2018) Agronomic and physiological response of giant reed (Arundo donax L.) to soil salinity. Ital J Agron 13:31–39. https://doi.org/10.4081/ija.2018.995
Evangelistella C, Valentini A, Ludovisi R, Firrincieli A, Fabbrini F, Scalabrin S, Cattonaro F, Morgante M, Mugnozza GS, Keurentjes JJB, Harfouche A (2017) De novo assembly, functional annotation, and analysis of the giant reed (Arundo donax L.) leaf transcriptome provide tools for the development of a biofuel feedstock. Biotechnol Biofuels 10(1):138
Ferreira LJ, Azevedo V, Maroco J et al (2015) Salt tolerant and sensitive rice varieties display differential methylome flexibility under salt stress. PLoS One 10(5):e0124060. https://doi.org/10.1371/journal.pone.0124060
Ferreira LJ, Donoghue MT, Barros P, Saibo NJ, Santos AP, Oliveira MM (2019) Uncovering Differentially Methylated Regions (DMRs) in a salt-tolerant rice variety under stress: one step towards new regulatory regions for enhanced salt tolerance. Epigenomes 3:4. https://doi.org/10.3390/epigenomes3010004
Fu Y, Poli M, Sablok G et al (2016) Dissection of early transcriptional responses to water stress in Arundo donax L. by unigene-based RNA-seq. Biotechnol Biofuels 9:54. https://doi.org/10.1186/s13068-016-0471-8
Fulneček J, Kovařík A (2014) How to interpret methylation sensitive amplified polymorphism (MSAP) profiles? BMC Genet 15:2. https://doi.org/10.1186/1471-2156-15-2
Galvan-Ampudia CS, Testerink C (2011) Salt stress signals shape the plant root. Curr Opin Plant Biol 14:296–302. https://doi.org/10.1016/J.PBI.2011.03.019
Garg R, Narayana Chevala V, Shankar R, Jain M (2015) Divergent DNA methylation patterns associated with gene expression in rice cultivars with contrasting drought and salinity stress response. Sci Rep 5:14922. https://doi.org/10.1038/srep14922
Gill S, Tuteja N (2010) Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant Physiol Biochem 48:909–930. https://doi.org/10.1016/j.plaphy.2010.08.016
Golldack D, Li C, Mohan H, Probst N (2014) Tolerance to drought and salt stress in plants: unraveling the signaling networks. Front Plant Sci 5:151. https://doi.org/10.3389/fpls.2014.00151
Goodstein DM, Shu S, Howson R, Neupane R, Hayes RD, Fazo J, Mitros T, Dirks W, Hellsten U, Putnam N, Rokhsar DS (2012) Phytozome: a comparative platform for green plant genomics. Nucl Acids Res 40:D1178–D1186. https://doi.org/10.1093/nar/gkr944
Guarino F, Cicatelli A, Brundu G, Improta G, Triassi M, Castiglione S (2019) The use of MSAP reveals epigenetic diversity of the invasive clonal populations of Arundo donax L. PLoS One 14:e0215096. https://doi.org/10.1371/journal.pone.0215096
Hasegawa PM, Bressan RA, Zhu J-K, Bohnert HJ (2000) Plant cellular and molecular responses to high salinity. Annu Rev Plant Physiol Plant Mol Biol 51:463–499. https://doi.org/10.1146/annurev.arplant.51.1.463
Haworth M, Catola S, Marino G et al (2017a) Moderate drought stress induces increased foliar dimethylsulphoniopropionate (DMSP) concentration and isoprene emission in two contrasting ecotypes of Arundo donax. Front Plant Sci 8:1016. https://doi.org/10.3389/fpls.2017.01016
Haworth M, Centritto M, Giovannelli A et al (2017b) Xylem morphology determines the drought response of two Arundo donax ecotypes from contrasting habitats. GCB Bioenergy 9:119–131. https://doi.org/10.1111/gcbb.12322
Haworth M, Cosentino SL, Marino G et al (2017c) Physiological responses of Arundo donax ecotypes to drought: a common garden study. GCB Bioenergy 9:132–143. https://doi.org/10.1111/gcbb.12348
Heath RL, Packer L (1968) Photoperoxidation in isolated chloroplasts: I. Kinetics and stoichiometry of fatty acid peroxidation. Arch Biochem Biophys 125:189–198. https://doi.org/10.1016/0003-9861(68)90654-1
Hoagland DR, Arnon DI (1950) The water-culture method for growing plants without soil. Circ Calif Agric Exp Stn 347:32 (citeulike-article-id:9455435)
Ismail A, Takeda S, Nick P (2014) Life and death under salt stress: same players, different timing? J Exp Bot 65:2963–2979. https://doi.org/10.1093/jxb/eru159
Julkowska MM, Testerink C (2015) Tuning plant signaling and growth to survive salt. Trends Plant Sci 20:586–594. https://doi.org/10.1016/j.tplants.2015.06.008
Karan R, DeLeon T, Biradar H, Subudhi PK (2012) Salt stress induced variation in DNA methylation pattern and its influence on gene expression in contrasting rice genotypes. PLoS One 7:e40203. https://doi.org/10.1371/journal.pone.0040203
Khan MS, Akther T, Hemalatha S (2017) Impact of Panchagavya on Oryza sativa L. grown under saline stress. J Plant Growth Regul 36:702–713. https://doi.org/10.1007/s00344-017-9674-x
Kobayashi NI, Yamaji N, Yamamoto H et al (2017) OsHKT1;5 mediates Na+exclusion in the vasculature to protect leaf blades and reproductive tissues from salt toxicity in rice. Plant J 91:657–670. https://doi.org/10.1111/tpj.13595
Lewandowski I, Scurlock JMO, Lindvall E, Christou M (2003) The development and current status of perennial rhizomatous grasses as energy crops in the US and Europe. Biomass Bioenergy 25:335–361
Lira-Medeiros CF, Parisod C, Fernandes RA, Mata CS, Cardoso MA, Ferreira PC (2010) Epigenetic variation in mangrove plants occurring in contrasting natural environment. PLoS One 5(4):e10326
Lízal P, Relichová J (2001) The effect of day length, vernalization and DNA demethylation on the flowering time in Arabidopsis thaliana. Physiol Plant 113:121–127. https://doi.org/10.1034/j.1399-3054.2001.1130116.x
Longstreth DJ, Nobel PS (1979) Salinity effects on leaf anatomy consequences for photosynthesis. Plant Physiol 63:700–703. https://doi.org/10.1104/pp.63.4.700
Maggio A, Raimondi G, Martino A, De Pascale S (2007) Salt stress response in tomato beyond the salinity tolerance threshold. Environ Exp Bot 59:276–282. https://doi.org/10.1016/j.envexpbot.2006.02.002
Mallikarjuna G, Mallikarjuna K, Reddy MK, Kaul T (2011) Expression of OsDREB2A transcription factor confers enhanced dehydration and salt stress tolerance in rice (Oryza sativa L.). Biotechnol Lett 33:1689–1697. https://doi.org/10.1007/s10529-011-0620-x
Malone JM, Virtue JG, Williams C, Preston C (2017) Genetic diversity of giant reed (Arundo donax) in Australia. Weed Biol Manag 17:17–28. https://doi.org/10.1111/wbm.12111
Marconi G, Pace R, Traini A et al (2013) Use of MSAP markers to analyse the effects of salt stress on DNA methylation in rapeseed (Brassica napus var. oleifera). PLoS One 8(9):e75597. https://doi.org/10.1371/journal.pone.0075597
Mariani C, Cabrini R, Danin A et al (2010) Origin, diffusion and reproduction of the giant reed (Arundo donax L.): a promising weedy energy crop. Ann Appl Biol 157:191–202. https://doi.org/10.1111/j.1744-7348.2010.00419.x
Miller G, Stein H, Honig A, Kapulnik Y, Zilberstein A (2005) Responsive modes of Medicago sativa proline dehydrogenase genes during salt stress and recovery dictate free proline accumulation. Planta 222(1):70–79
Mittler R (2002) Oxidative stress, antioxidants and stress tolerance. Trends Plant Sci 7:405–410
Moller IS, Gilliham M, Jha D et al (2009) Shoot Na+ exclusion and increased salinity tolerance engineered by cell type-specific alteration of Na+ transport in Arabidopsis. Plant Cell 21:2163–2178. https://doi.org/10.1105/tpc.108.064568
Moran JF, Becana M, Iturbe-Ormaetxe I et al (1994) Drought induces oxidative stress in pea plants. Planta 194:346–352
Munns R, Tester M (2008) Mechanisms of salinity tolerance. Annu Rev Plant Biol 59:651–681. https://doi.org/10.1146/annurev.arplant.59.032607.092911
Nackley LL, Kim SH (2015) A salt on the bioenergy and biological invasions debate: salinity tolerance of the invasive biomass feedstock Arundo donax. GCB Bioenergy 7:752–762. https://doi.org/10.1111/gcbb.12184
Negrao S, Schmöckel SM, Tester M et al (2017) Evaluating physiological responses of plants to salinity stress. Ann Bot 119:1–11. https://doi.org/10.1093/aob/mcw191
Papazoglou EG (2007) Arundo donax L. stress tolerance under irrigation with heavy metal aqueous solutions. Desalination 211:304–313. https://doi.org/10.1016/j.desal.2006.03.600
Pfaffl MW (2001) A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29:e45. https://doi.org/10.1093/nar/29.9.e45
Pfaffl MW (2004) Quantification strategies in real-time PCR. In: Bustin SA (ed) A-Z of quantitative PCR 87–112. International University Line (IUL), La Jolla. https://doi.org/10.1007/s10551-011-0963-1
Pilu R, Cassani E, Landoni M et al (2014) Genetic characterization of an Italian giant reed (Arundo donax L.) clones collection: exploiting clonal selection. Euphytica 196:169–181. https://doi.org/10.1007/s10681-013-1022-z
Poli M, Salvi S, Li M, Varotto C (2017) Selection of reference genes suitable for normalization of qPCR data under abiotic stresses in bioenergy crop Arundo donax L. Sci Rep 7(1):10719. https://doi.org/10.1038/s41598-017-11019-0
Pollastri S, Savvides A, Pesando M et al (2017) Impact of two arbuscular mycorrhizal fungi on Arundo donax L. response to salt stress. Planta 247:573–585. https://doi.org/10.1007/s00425-017-2808-3
Pompeiano A, Landi M, Meloni G et al (2017) Allocation pattern, ion partitioning, and chlorophyll a fluorescence in Arundo donax L. in responses to salinity stress. Plant Biosyst 151(4):613–622. https://doi.org/10.1080/11263504.2016.1187680
Price AH, Atherton NM, Hendry GAF (1989) Plants under drought-stress generate activated oxygen. Free Radic Res 8:61–66. https://doi.org/10.3109/10715768909087973
Qiu Q-S, Guo Y, Dietrich MA et al (2002) Regulation of SOS1, a plasma membrane Na+/H+ exchanger in Arabidopsis thaliana, by SOS2 and SOS3. Proc Natl Acad Sci USA 99:8436–8441. https://doi.org/10.1073/pnas.122224699
Sablok G, Fu Y, Bobbio V, Laura M, Rotino GL, Bagnaresi P, Allavena A, Velikova V, Viola R, Loreto F, Li M, Varotto C (2014) Fuelling genetic and metabolic exploration of C3 bioenergy crops through the first reference transcriptome of Arundo donax L. Plant Biotechnol J 12(5):554–567
Sairam RK, Tyagi A (2004) Physiology and molecular biology of salinity stress tolerance in plants. Curr Sci 86:407–421. https://doi.org/10.1016/j.tplants.2005.10.002
Sakuma Y, Maruyama K, Osakabe Y et al (2006) Functional analysis of an Arabidopsis transcription factor, DREB2A, involved in drought-responsive gene expression. Plant Cell 18:1292–1309. https://doi.org/10.1105/tpc.105.035881.1
Saltonstall K, Lambert A, Meyerson LA (2010) Genetics and reproduction of common (Phragmites australis) and giant reed (Arundo donax). Invasive Plant Sci Manag 3:495–505. https://doi.org/10.1614/IPSM-09-053.1
Sánchez E, Scordia D, Lino G et al (2015) Salinity and water stress effects on biomass production in different Arundo donax L. clones. BioEneregy Res 8(4):1461–1479. https://doi.org/10.1007/s12155-015-9652-8
Schulz B, Eckstein RL, Durka W (2013) Scoring and analysis of methylation-sensitive amplification polymorphisms for epigenetic population studies. Mol Ecol Resour 13:642–653. https://doi.org/10.1111/1755-0998.12100
Spencer DF (2014) Evaluation of stem injection for managing giant reed (Arundo donax). J Environ Sci Health Part B 49(9):633–638
Song X, Wang J, Ma X et al (2016) Origination, expansion, evolutionary trajectory, and expression bias of AP2/ERF superfamily in Brassica napus. Front Plant Sci 7:1186. https://doi.org/10.3389/fpls.2016.01186
Sun Y, Yu D (2015) Activated expression of AtWRKY53 negatively regulates drought tolerance by mediating stomatal movement. Plant Cell Rep 34:1295–1306. https://doi.org/10.1007/s00299-015-1787-8
Tang AC, Boyer JS (2007) Leaf shrinkage decreases porosity at low water potentials in sunflower. Funct Plant Biol 34:24–30. https://doi.org/10.1071/FP06222
Van Eck L, Davidson RM, Wu S et al (2014) The transcriptional network of WRKY53 in cereals links oxidative responses to biotic and abiotic stress inputs. Funct Integr Genomics 14:351–362. https://doi.org/10.1007/s10142-014-0374-3
Wang W, Huang F, Qin Q et al (2015) Comparative analysis of DNA methylation changes in two rice genotypes under salt stress and subsequent recovery. Biochem Biophys Res Commun 465:790–796. https://doi.org/10.1016/j.bbrc.2015.08.089
Yang Q, Chen ZZ, Zhou XF et al (2009) Overexpression of SOS (salt overly sensitive) genes increases salt tolerance in transgenic Arabidopsis. Mol Plant 2:22–31. https://doi.org/10.1093/mp/ssn058
Yoshimura K (2008) Programmed proteome response for drought avoidance/tolerance in the root of a C3 xerophyte (wild watermelon) under water deficits. Plant Cell Physiol 49:226–241. https://doi.org/10.1093/pcp/pcm180
Zhang XX, Tang YJ, Bin Ma Q et al (2013) OsDREB2A, a rice transcription factor, significantly affects salt tolerance in transgenic soybean. PLoS One 8(12):e83011. https://doi.org/10.1371/journal.pone.0083011
Zhang H, Lang Z, Zhu JK (2018) Dynamics and function of DNA methylation in plants. Nat Rev Mol Cell Biol 19:489–506. https://doi.org/10.1038/s41580-018-0016-z
Zhu J (2016) Abiotic stress signaling and responses in plants. Cell 167:313–324. https://doi.org/10.1016/j.cell.2016.08.029
Acknowledgements
We are thankful to Prof. G. Caruso (Università di Napoli Federico II, Italy) for valuable help with statistical analysis. Assistance of Mr. G. Guarino and Mr. R. Nocerino (CNR-IBBR, Portici, Italy) with artworks and plant growth, respectively, is gratefully acknowledged. This work was partially supported by a research grant from the Italian Ministry of Education, University and Research, Project BioPoliS (PON03PE_00107_1).
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Docimo, T., De Stefano, R., De Palma, M. et al. Transcriptional, metabolic and DNA methylation changes underpinning the response of Arundo donax ecotypes to NaCl excess. Planta 251, 34 (2020). https://doi.org/10.1007/s00425-019-03325-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00425-019-03325-w