Abstract
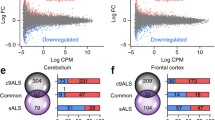
We previously found C9orf72-associated (c9ALS) and sporadic amyotrophic lateral sclerosis (sALS) brain transcriptomes comprise thousands of defects, among which, some are likely key contributors to ALS pathogenesis. We have now generated complementary methylome data and combine these two data sets to perform a comprehensive “multi-omic” analysis to clarify the molecular mechanisms initiating RNA misregulation in ALS. We found that c9ALS and sALS patients have generally distinct but overlapping methylome profiles, and that the c9ALS- and sALS-affected genes and pathways have similar biological functions, indicating conserved pathobiology in disease. Our results strongly implicate SERPINA1 in both C9orf72 repeat expansion carriers and non-carriers, where expression levels are greatly increased in both patient groups across the frontal cortex and cerebellum. SERPINA1 expression is particularly pronounced in C9orf72 repeat expansion carriers for both brain regions, where SERPINA1 levels are strictly down regulated across most human tissues, including the brain, except liver and blood, and are not measurable in E18 mouse brain. The altered biological networks we identified contain critical molecular players known to contribute to ALS pathology, which also interact with SERPINA1. Our comprehensive combined methylation and transcription study identifies new genes and highlights that direct genetic and epigenetic changes contribute to c9ALS and sALS pathogenesis.




Similar content being viewed by others
References
Akalin A, Kormaksson M, Li S, Garrett-Bakelman FE, Figueroa ME, Melnick A et al (2012) methylKit: a comprehensive R package for the analysis of genome-wide DNA methylation profiles. Genome Biol 13:R87. doi:10.1186/gb-2012-13-10-r87
Amaral PP, Dinger ME, Mercer TR, Mattick JS (2008) The eukaryotic genome as an RNA machine. Science 319:1787–1789. doi:10.1126/science.1155472
Belzil VV, Bauer PO, Gendron TF, Murray ME, Dickson D, Petrucelli L (2014) Characterization of DNA hypermethylation in the cerebellum of c9FTD/ALS patients. Brain Res 1584:15–21. doi:10.1016/j.brainres.2014.02.015
Belzil VV, Bauer PO, Prudencio M, Gendron TF, Stetler CT, Yan IK et al (2013) Reduced C9orf72 gene expression in c9FTD/ALS is caused by histone trimethylation, an epigenetic event detectable in blood. Acta Neuropathol 126:895–905. doi:10.1007/s00401-013-1199-1
Belzil VV, Gendron TF, Petrucelli L (2012) RNA-mediated toxicity in neurodegenerative disease. Mol Cell Neurosci 56C:406–419. doi:10.1016/j.mcn.2012.12.006
Belzil VV, Katzman RB, Petrucelli L (2016) ALS and FTD: an epigenetic perspective. Acta Neuropathol. doi:10.1007/s00401-016-1587-4
Byrne S, Heverin M, Elamin M, Bede P, Lynch C, Kenna K et al (2013) Aggregation of neurologic and neuropsychiatric disease in amyotrophic lateral sclerosis kindreds: a population-based case-control cohort study of familial and sporadic amyotrophic lateral sclerosis. Ann Neurol 74:699–708. doi:10.1002/ana.23969
Byrne S, Walsh C, Lynch C, Bede P, Elamin M, Kenna K et al (2011) Rate of familial amyotrophic lateral sclerosis: a systematic review and meta-analysis. J Neurol Neurosurg Psychiatry 82:623–627. doi:10.1136/jnnp.2010.224501
Cox DW (2001) Alpha-1-antitrypsin. In: Scriver CRB, Sly AL, Valle D (eds) The metabolic and molecular bases of inherited disease, vol IV, 8th edn. McGraw-Hill, New York, pp 5559–5584
Donnelly CJ, Zhang PW, Pham JT, Haeusler AR, Mistry NA, Vidensky S et al (2013) RNA toxicity from the ALS/FTD C9ORF72 expansion is mitigated by antisense intervention. Neuron 80:415–428. doi:10.1016/j.neuron.2013.10.015
Fratta P, Mizielinska S, Nicoll AJ, Zloh M, Fisher EM, Parkinson G et al (2012) C9orf72 hexanucleotide repeat associated with amyotrophic lateral sclerosis and frontotemporal dementia forms RNA G-quadruplexes. Sci Rep 2:1016. doi:10.1038/srep01016
Gendron TF, van Blitterswijk M, Bieniek KF, Daughrity LM, Jiang J, Rush BK et al (2015) Cerebellar c9RAN proteins associate with clinical and neuropathological characteristics of C9ORF72 repeat expansion carriers. Acta Neuropathol 130:559–573. doi:10.1007/s00401-015-1474-4
Gibson SB, Figueroa KP, Bromberg MB, Pulst SM, Cannon-Albright L (2014) Familial clustering of ALS in a population-based resource. Neurology 82:17–22. doi:10.1212/01.wnl.0000438219.39061.da
Guerreiro R, Bras J, Hardy J (2015) SnapShot: genetics of ALS and FTD. Cell 160(798):e791. doi:10.1016/j.cell.2015.01.052
Haeusler AR, Donnelly CJ, Periz G, Simko EA, Shaw PG, Kim MS et al (2014) C9orf72 nucleotide repeat structures initiate molecular cascades of disease. Nature 507:195–200. doi:10.1038/nature13124
Holoch D, Moazed D (2015) RNA-mediated epigenetic regulation of gene expression. Nat Rev Genet 16:71–84. doi:10.1038/nrg3863
Jung J, Bonini N (2007) CREB-binding protein modulates repeat instability in a Drosophila model for polyQ disease. Science 315:1857–1859. doi:10.1126/science.1139517
Kalsheker NA (1996) Alpha 1-antichymotrypsin. Int J Biochem Cell Biol 28:961–964
Kamboh MI, Minster RL, Kenney M, Ozturk A, Desai PP, Kammerer CM et al (2006) Alpha-1-antichymotrypsin (ACT or SERPINA3) polymorphism may affect age-at-onset and disease duration of Alzheimer’s disease. Neurobiol Aging 27:1435–1439. doi:10.1016/j.neurobiolaging.2005.07.015
Krueger F, Andrews SR (2011) Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics 27:1571–1572. doi:10.1093/bioinformatics/btr167
Lam L, Chin L, Halder RC, Sagong B, Famenini S, Sayre J et al (2016) Epigenetic changes in T-cell and monocyte signatures and production of neurotoxic cytokines in ALS patients. FASEB J. doi:10.1096/fj.201600259RR
Langmead B, Salzberg SL (2012) Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–359. doi:10.1038/nmeth.1923
Liu EY, Russ J, Wu K, Neal D, Suh E, McNally AG et al (2014) C9orf72 hypermethylation protects against repeat expansion-associated pathology in ALS/FTD. Acta Neuropathol 128:525–541. doi:10.1007/s00401-014-1286-y
Mattick JS, Amaral PP, Dinger ME, Mercer TR, Mehler MF (2009) RNA regulation of epigenetic processes. BioEssays 31:51–59. doi:10.1002/bies.080099
Morahan JM, Yu B, Trent RJ, Pamphlett R (2009) A genome-wide analysis of brain DNA methylation identifies new candidate genes for sporadic amyotrophic lateral sclerosis. Amyotroph Lateral Scler 10:418–429. doi:10.3109/17482960802635397
Morris KV, Mattick JS (2014) The rise of regulatory RNA. Nat Rev Genet 15:423–437. doi:10.1038/nrg3722
Nagase T, Nakayama M, Nakajima D, Kikuno R, Ohara O (2001) Prediction of the coding sequences of unidentified human genes. XX. The complete sequences of 100 new cDNA clones from brain which code for large proteins in vitro. DNA Res 8:85–95
Neumann M, Sampathu DM, Kwong LK, Truax AC, Micsenyi MC, Chou TT et al (2006) Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science 314:130–133
Pace JM, Corrado M, Missero C, Byers PH (2003) Identification, characterization and expression analysis of a new fibrillar collagen gene, COL27A1. Matrix Biol 22:3–14
Pennacchio LA, Bickmore W, Dean A, Nobrega MA, Bejerano G (2013) Enhancers: five essential questions. Nat Rev Genet 14:288–295. doi:10.1038/nrg3458
Polymenidou M, Lagier-Tourenne C, Hutt KR, Bennett CF, Cleveland DW, Yeo GW (2012) Misregulated RNA processing in amyotrophic lateral sclerosis. Brain Res 1462:3–15. doi:10.1016/j.brainres.2012.02.059
Prell T, Grosskreutz J (2013) The involvement of the cerebellum in amyotrophic lateral sclerosis. Amyotroph Lateral Scler Frontotemporal Degener 14:507–515. doi:10.3109/21678421.2013.812661
Prudencio M, Belzil VV, Batra R, Ross CA, Gendron TF, Pregent LJ et al (2015) Distinct brain transcriptome profiles in C9orf72-associated and sporadic ALS. Nat Neurosci 18:1175–1182. doi:10.1038/nn.4065
Rademakers R, van Blitterswijk M (2013) Motor neuron disease in 2012: novel causal genes and disease modifiers. Nat Rev Neurol 9:63–64. doi:10.1038/nrneurol.2012.276
Renoux AJ, Todd PK (2012) Neurodegeneration the RNA way. Prog Neurobiol 97:173–189. doi:10.1016/j.pneurobio.2011.10.006
Renton AE, Chio A, Traynor BJ (2014) State of play in amyotrophic lateral sclerosis genetics. Nat Neurosci 17:17–23. doi:10.1038/nn.3584
Spitz F (2016) Gene regulation at a distance: from remote enhancers to 3D regulatory ensembles. Semin Cell Dev Biol. doi:10.1016/j.semcdb.2016.06.017
Su Z, Zhang Y, Gendron TF, Bauer PO, Chew J, Yang WY et al (2014) Discovery of a biomarker and lead small molecules to target r(GGGGCC)-associated defects in c9FTD/ALS. Neuron 83:1043–1050. doi:10.1016/j.neuron.2014.07.041
Tan RH, Kril JJ, McGinley C, Hassani M, Masuda-Suzukake M, Hasegawa M et al (2016) Cerebellar neuronal loss in amyotrophic lateral sclerosis cases with ATXN2 intermediate repeat expansions. Ann Neurol 79:295–305. doi:10.1002/ana.24565
Therrien M, Dion PA, Rouleau GA (2016) ALS: recent Developments from Genetics Studies. Curr Neurol Neurosci Rep 16:59. doi:10.1007/s11910-016-0658-1
Wang YC, Liu HC, Liu TY, Hong CJ, Tsai SJ (2001) Genetic association analysis of alpha-1-antichymotrypsin polymorphism in Parkinson’s disease. Eur Neurol 45:254–256. doi:10.1159/000052138
Xi Z, Rainero I, Rubino E, Pinessi L, Bruni AC, Maletta RG et al (2014) Hypermethylation of the CpG-island near the C9orf72 G4C2-repeat expansion in FTLD patients. Hum Mol Genet. doi:10.1093/hmg/ddu279
Xi Z, Yunusova Y, van Blitterswijk M, Dib S, Ghani M, Moreno D et al (2014) Identical twins with the C9orf72 repeat expansion are discordant for ALS. Neurology 83:1476–1478. doi:10.1212/WNL.0000000000000886
Xi Z, Zhang M, Bruni AC, Maletta RG, Colao R, Fratta P et al (2015) The C9orf72 repeat expansion itself is methylated in ALS and FTLD patients. Acta Neuropathol 129:715–727. doi:10.1007/s00401-015-1401-8
Xi Z, Zinman L, Moreno D, Schymick J, Liang Y, Sato C et al (2013) Hypermethylation of the CpG island near the G4C2 repeat in ALS with a C9orf72 expansion. Am J Hum Genet 92:981–989. doi:10.1016/j.ajhg.2013.04.017
Zeng Q, Subramaniam VN, Wong SH, Tang BL, Parton RG, Rea S et al (1998) A novel synaptobrevin/VAMP homologous protein (VAMP5) is increased during in vitro myogenesis and present in the plasma membrane. Mol Biol Cell 9:2423–2437
Zhang K, Donnelly CJ, Haeusler AR, Grima JC, Machamer JB, Steinwald P et al (2015) The C9orf72 repeat expansion disrupts nucleocytoplasmic transport. Nature 525:56–61. doi:10.1038/nature14973
Acknowledgements
We are extremely grateful to all individuals who agreed to donate their brains to research. This study was supported by the National Institutes of Health/National Institute on Aging [AG16574-17 J PILOT (V.V.B.)]; National Institutes of Health/National Institute of Neurological Disorders and Stroke [R21NS074121 (K.B.B.), P01NS084974 (D.W.D., K.B.B., and R.R.]; Mayo Clinic Center for Individualized Medicine (V.V.B., K.B.B., and H.L.); ALS Association (K.B.B.), Donors Cure Foundation (H.L.), and the Robert Packard Center for ALS Research at Johns Hopkins. V.V.B. is recipient of the Career Transition Award from ALS Canada and Brain Canada, the Milton Safenowitz Post-Doctoral Fellowship from the Amyotrophic Lateral Sclerosis Association, the Post-Doctoral Fellowship from the Canadian Institutes of Health Research, the Career Development Award for Young Investigators in Neurosciences from the Siragusa Foundation, and the Research Fellowship from the Robert and Clarice Smith & Abigail Van Buren Alzheimer’s Disease Research Foundation. M.T.W.E received the PhRMA Foundation Research Starter grant.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Ebbert, M.T.W., Ross, C.A., Pregent, L.J. et al. Conserved DNA methylation combined with differential frontal cortex and cerebellar expression distinguishes C9orf72-associated and sporadic ALS, and implicates SERPINA1 in disease. Acta Neuropathol 134, 715–728 (2017). https://doi.org/10.1007/s00401-017-1760-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00401-017-1760-4