Abstract
Metabolic modulation is a promising therapeutic approach to prevent adverse remodeling of the ischemic heart. Because little is known about the involvement of long non-coding RNAs (lncRNAs) in regulating cardiac metabolism, we used unbiased transcriptome profiling in a mouse model of myocardial infarction (MI). We identified a novel cardiomyocyte-enriched lncRNA, called LncHrt, which regulates metabolism and the pathophysiological processes that lead to heart failure. AAV-based LncHrt overexpression protects the heart from MI as demonstrated by improved contractile function, preserved metabolic homeostasis, and attenuated maladaptive remodeling responses. RNA-pull down followed by mass spectrometry and RNA immunoprecipitation (RIP) identified SIRT2 as a LncHrt-interacting protein involved in cardiac metabolic regulation. Mechanistically, we established that LncHrt interacts with SIRT2 to preserve SIRT2 deacetylase activity by interfering with the CDK5 and SIRT2 interaction. This increases downstream LKB1-AMPK kinase signaling, which ameliorates functional and metabolic deficits. Importantly, we found the expression of the human homolog of mouse LncHrt was decreased in patients with dilated cardiomyopathy. Together, these studies identify LncHrt as a cardiac metabolic regulator that plays an essential role in preserving heart function by regulating downstream metabolic signaling pathways. Consequently, LncHrt is a potentially novel RNA-based therapeutic target for ischemic heart disease.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Myocardial infarction (MI) is the leading cause of global mortality and morbidity [51] and typically occurs because of an acute blockage in a coronary artery. The ensuing tissue ischemia can cause cardiomyopathy, which is characterized by contractile dysfunction, scar formation, and structural remodeling [7, 74]. Cardiac remodeling responses to ischemia are initially functional, compensatory, and adaptive; but, when sustained, structural changes extend well beyond the region of injury, which leads to pathological hypertrophy accompanied by metabolic changes, including decreased fatty acid oxidation and increased glycolysis [19]. These effects decrease ATP synthesis, which, in turn, contributes to increased susceptibility of the injured heart to the progression toward failure [14, 19, 28, 45, 46]. Therefore, optimizing metabolic remodeling responses following myocardial infarction may provide an innovative therapeutic opportunity [19].
The mammalian heart is a ‘metabolic omnivore’ that prefers fatty acids as the fuel for oxidative phosphorylation. This preference can change based on environmental stress and is regulated at the level of enzyme activity as well as transcription, translation, and post-translational modification of proteins [22, 25]. During an ischemic episode, the activation of AMP-activated protein kinase (AMPK) is an adaptive response to cardiac energy deficit that prolongs the survival and function of cardiomyocytes. AMPK activation conserves energy by phosphorylating downstream targets that inhibit ATP-consuming anabolic processes and promote ATP-generating catabolic processes. By modulating cardiac metabolic homeostasis, AMPK activity decreases cardiomyocyte death and improves cardiac function [26, 52, 57]. Despite extensive study of AMPK and the upstream kinase LKB1 in regulating cardiac metabolic homeostasis, the role of long noncoding RNAs (lncRNAs) in these processes remain unclear.
Transcriptome analyses show thousands of lncRNAs are differentially expressed during heart development and disease [32, 47]. LncRNAs act as key modulators of gene expression that act epigenetically, post-transcriptionally, and translationally. Emerging evidence has shown that the manipulation of some lncRNAs can ameliorate dysfunction of the heart by regulating cardiac regeneration, hypertrophy, fibrosis, and angiogenesis [3, 43, 62, 64]. Our previous study showed that lincRNA-p21 plays a critical role in cell proliferation and neointimal formation during cardiovascular remodeling [68]. We also reported that many lncRNAs are dynamically expressed in ischemic cardiomyopathy patient samples, which results in extracellular matrix changes [20].
Using unbiased-screening of heart tissue 3 and 14 days after MI, we have previously reported that the expression of a cardiac fibroblast-enriched lncRNA, Cfast, was significantly induced in the heart of MI. Inhibition of Cfast preserved cardiac function by repressing fibrotic remodeling following cardiac injury [73]. Here, we report the identification of a cardiomyocyte-enriched lncRNA, named LncHrt, that is dramatically downregulated in infarcted hearts. We show LncHrt is essential for maintaining cardiac homeostasis and is sufficient to positively regulate cardiac metabolism and improve function post-MI. Mechanistically, LncHrt preserves cardiac metabolic balance through an interaction with SIRT2, which preserves its activity by interfering with the interaction between CDK5 and SIRT2. This abrogates the suppressive role CDK5 has on SIRT2 catalytic activity and activates downstream signaling of the LKB1-AMPK cascades to protect the heart from adverse remodeling responses.
Materials and methods
Animals
Experiments involving animals were performed in accordance with the Guide for the Care and Use of Laboratory Animals and were approved by the Institutional Animal Care and Use Committee (IACUC) of Boston Children’s Hospital (BCH) and by the IACUC of Zhejiang University. The animals were fed a standard laboratory diet and maintained with a 12:12 h light/dark cycle.
Human tissue samples
Left ventricular (LV) and aorta tissues were taken from patients with dilated cardiomyopathy during heart transplantation. Surgeries were performed between September 2014 and February 2019. In brief, the patient’s heart was removed at the time of transplantation, LV and aorta tissues were dissected and snap frozen for subsequent analysis. LV samples from donated healthy hearts were used as a control. All subjects were duly informed and gave written consent. All studies were approved by the Ethics Review Committee of Second Affiliated Hospital of Zhejiang University.
Microarray-based lncRNA profiling
Total RNAs were prepared from the heart tissues of mice 3 and 14 days after myocardial infarction (MI) (n = 5 hearts per group) or sham surgery (n = 4 hearts) (see below). The RNA samples were used for global lncRNA profiling with the LncRNA Microarray SurePrint G3 Gene Mouse GE 8 × 60K Kit (Agilent Technologies, Inc.) according to the manufacturer’s protocols. Agilent Feature Extraction software (version 11.0.1.1) was used to analyze acquired array images. Quantile-normalization and subsequent data processing were performed with the GeneSpring GX v12.1 software package (Agilent Technologies, Inc.). Differentially expressed LncRNAs were identified through filtering on Fold Change (≥ 2) and P value (p < 0.05).
Quantitative reverse transcription-polymerase chain reaction (qRT-PCR)
Total RNA was isolated using TRIzol reagent (Life Technologies) from heart tissues and incubated with DNase I (Life technologies) to remove residual genomic DNA. For detecting RNA levels with qRT-PCR, one microgram RNA was used to synthesize cDNA following the manufacturer’s instructions (TAKARA). qRT-PCR signal was detected by the VII7 Real-time PCR System with a SYBR green qPCR Master Mix (Vazyme Biotech). ddCt was used to analyze gene expression from qPCR datasets. Data were normalized to 18S rRNA (RN18s) or Actin-beta (Actb) signal. Primers used for qRT-PCR in this study are listed in Supplementary Table 10.
Isolation of adult mouse cardiac cells
Adult mouse cardiomyocyte and non-cardiomyocyte isolations were performed on 2- to 3-month-old mice using a previously reported procedure[5]. Briefly, the mouse heart was perfused and digested with collagenase II (Worthington Biochemical Corp, Lakewood, NJ), and dissociated cells were sedimented by gravity. The supernatant, enriched in non-cardiomyocytes, was collected by centrifuging for 5 min at 1000 rpm. The bottom layer, rich in cardiomyocytes, also was collected. The adult non-cardiomyocytes and cardiomyocytes were then treated with Trizol for RNA extraction and qRT-PCR analyses.
RNA fractionation
Nuclear and cytoplasmic RNA fractions of cardiomyocytes were isolated using NE-PER Nuclear and Cytoplasmic Extraction Reagents (Thermo Scientific) according to the manufacturer’s instructions with addition of RNase inhibitor. The extracted nuclear and cytoplasmic RNA fractions were used to detect LncHrt subcellular content by qRT-PCR. The expression of Actb was used as a cytoplasmic control, and U6 was used as a nuclear control.
Rapid amplification of cDNA ends (RACE)
The full-length cDNA sequence of LncHrt was obtained from mouse heart RNA according to the SMARTer RACE 5ʹ/3ʹ Kit User Manual (TaKaRa). Nested 5ʹ and 3ʹ RACE products were obtained using GXL Taq polymerase with GC Buffer (Takara). The primers used for RACE PCR and nested PCR are presented in Supplementary Table 12. PCR products were extracted with a Gel Extraction kit (Vazyme Biotech), cloned into the pMD18-T vector, and analyzed by Sanger sequencing.
AAV9 cloning, virus packaging and injection
HEK293T (TAKARA) cells were maintained in Dulbecco’s Modified Eagle’s Medium (DMEM, Sigma) supplemented with 10% fetal bovine serum (FBS; Gibco) and penicillin (100 U/mL)/streptomycin (100 µg/mL) (Life Technologies).
AAV9-cTNT:LncHrt (AAV-LncHrt) was generated in the AAV9 vector where the cardiac-specific TNNT2 promoter was used to drive the expression of LncHrt in the heart. An AAV9-cTNT:Luciferase (AAV-CTRL) was used as a control. The cDNA fragments encoding Luciferase and LncHrt were separately cloned into the AAV Inverted Terminal Repeat (ITR)-containing AAV9 plasmid (Penn Vector Core) harboring the chicken cardiac TNT promoter, to generate pEn.cTnT::Luciferase and pEn.cTnT::LncHrt, respectively. The AAV9-U6::LncHrt shRNA was generated using the pCRII Topo U6 vector, where the U6 promoter was used to drive the expression of the target LncHrt shRNA sequence in the heart to silencing LncHrt expression. The targeting sites of shRNA for LncHrt were designed using SirnaWizard (https://www.invivogen.com/sirnawizard/design.php), which are provide in Supplementary Table 7.
The AAV9 vectors were packaged in HEK293T cells with AAV9:Rep-Cap and pAd deltaF6 (Penn Vector Core) as described previously [15]. Transfections were performed using 1 mg/mL polyethylenimine, branched (Sigma) using a 1:4 dilution. After 60 h, cells were collected and lysed. AAV was purified and concentrated by gradient centrifugation. Titration of AAV particles was achieved using qRT- PCR, as described previously [12, 21]. The viral preparations had titers between 2 × 1013 and 5 × 1013 viral genome particles per mL. For AAV injection, 1-day-old neonatal C57BL/6 mice were randomly subjected to subcutaneous injection of AAV-LncHrt/AAV-LncHrt KD or AAV9-Control/Scramble (2 ~ 5 × 1011 viral genome particles per mouse heart), respectively; when mice were 2 months old, they were used to detect cardiac function or used for MI surgery. Intra-myocardial injections of AAV9-LncHrt or AAV9-Control (2 ~ 5 × 1011 viral genome particles per mouse heart), respectively, post-MI were performed as per our previous report [12]. Immediately after the ligation of LAD coronary artery, AAV9-LncHrt or AAV9-Control in a total volume of 30 μL was injected into ventricle muscular wall but not the ventricular cavity. The AAV9-LncHrt or AAV9-Control was evenly injected into four sites around the infarcted area (two sites in anterior wall, lateral wall, and apex area). Then, the chest was closed and the animal was placed in a prone position until recovery of spontaneous breathing.
In vitro Cdk5 and Sirt2 silencing
Neonatal mouse cardiomyocytes (NMCMs) were isolated from 1- to 2-day-old C57BL/6 mice by enzymatic disassociation according to the manual of neonatal heart dissociation Kit (Miltenyi Biotec, Germany). Isolated cardiomyocytes were grown in M199 Medium containing 10% fetal bovine serum (FBS, Gibco) and Penicillin–Streptomycin (100 U/mL, Gibco) and 20 µM cytosine β-D-arabinofuranoside (Sigma). After 16–18 h, NMCMs were cultured in serum-free M199 for 24 h, then treated with siRNAs targeting Cdk5 (Gene Pharma Corp). Targeting siRNAs and scramble siRNAs were transfected into NMCMs at a final concentration of 50 nM using Lipofectamine RNAiMAX Reagent (Thermo). The NMCMs were also treated with 5 µM Sirt2 inhibitor sirReal2 (Selleck).
Myocardial infarction
Myocardial infarction surgery was performed on 2-month-old C57BL/6 mice by ligation of the left anterior descending (LAD) coronary artery [60]. Mice were anesthetized with isoflurane. After the chest was shaved and cleaned with 75% alcohol, a suture was placed around the front upper incisors and pulled taut so that the neck was slightly extended. For oral intubation, the tongue was retracted and held with forceps, and a 20G catheter was inserted into the trachea. The catheter was then attached to the mouse ventilator via a Y-shaped connector. Ventilation was performed with a tidal volume of 225 µL for a 25-g mouse and a respiratory rate of 130 breaths per minute. 100% oxygen was provided to the inflow of the ventilator. Then the chest was opened through a left parasternal incision, and the heart exposed at the left 3rd–4th intercostal space. Chest retractor was applied to facilitate the view. The pericardium was opened, and the ligation was performed on the LAD coronary artery using 7–0 silk sutures (Ethicon). The lungs were slightly overinflated to assist in removal of air in the pleural cavity. Dissected intercostal space and chest skin were closed using a 6–0 silk suture (Ethicon). The sham group underwent the same surgical procedure (except that the LCA was not occluded).
Measurement of cardiac function by echocardiography
Echocardiographic measurements were performed on mice using a Visual Sonics Vevo 2100 Imaging System (Visual Sonics, Toronto, Canada) with a 18–38 MHz transducer (model MS-400). Left ventricular (LV) dimensions, including diastolic and systolic wall thicknesses, LV end-diastolic and end-systolic chamber dimensions were measured from 2-D short-axis under M-mode tracings at the level of the papillary muscle. LV mass and functional parameters such as percentage of fractional shortening (FS%) and left ventricular volume were calculated using the above primary measurements and accompanying software.
Two-dimensional M-mode images of the left ventricle were captured at the level of the papillary muscles, Left ventricular (LV) dimensions, including diastolic (LVPW;d) and systolic (LVPW;s) wall thickness, and the left ventricular internal diameter at end-diastole (LVID;d) and end-systole (LVID;s) were measured. Percentage ejection fraction (EF) and percentage fractional shortening (FS) were calculated as described previously [70].
Histology and immunostaining
Mouse hearts were dissected out, rinsed, and arrested in diastole buffer (4.7 nM KCl and 0.1% BDM in PBS). Hearts were then fixed in 4% paraformaldehyde (pH 7.4) overnight. After dehydration through a series of ethanol baths, samples were embedded in paraffin wax according to standard laboratory procedures. Sections of 4 µm in thickness were stained with hematoxylin and eosin (H&E). The stained sections were used for histological examination by light microscopy. To determine infarct size, the embedded paraffin blocks were cut through from apex to base. The first 5 sections of every 200 sections were used to stain with Fast Green and Sirius Red. Infarct size was calculated according to the following formula: [length of coronal infarct perimeter (epicardial + endocardial)/total left ventricular coronal perimeter (epicardial + endocardial)] × 100 [12, 50].
Immunofluorescence staining was performed on 4% paraformaldehyde-fixed and paraffin-embedded heart sections to measure the cardiomyocytes areas. After deparaffinization, rehydration, and heat-induced antigen retrieval, sections were incubated with wheat germ agglutinin (Alexa Fluor® 647 conjugate WGA; 1:500, Invitrogen) for labeling the cardiomyocyte membrane. Image J software was used to measure the size of cardiomyocytes in the papillary muscles.
RNA-seq and genome-wide transcriptome analysis
Total RNAs from the heart tissues of AAV-LncHrt and AAV-CTRL injected mice 7 days after MI was prepared for RNA-seq (three biological replicates for each group). RNA-seq experiments were performed by Novogene (Beijing, China). Briefly, total RNA was isolated from fresh ventricular tissue using TRIzol (Invitrogen). mRNA was then purified from total RNA using poly-T oligo-attached magnetic beads. Sequencing libraries were generated using NEBNext® UltraTM RNA Library Prep Kit for Illumina® (NEB, USA) following the manufacturer’s recommendations, and index codes were added to attribute sequences to each sample.
The clustering of the index-coded samples was performed on a cBot Cluster Generation System using TruSeq PE Cluster Kit v3-cBot-HS (Illuminia) according to the manufacturer’s instructions. After cluster generation, the library preparations were sequenced on an Illumina Hiseq platform and 150 bp paired-end reads were generated. For the data analysis, raw data (raw reads) in fastq format were first processed through in-house Perl scripts. Clean data (clean reads) were obtained by removing reads containing adapters, reads containing ploy-N, and low-quality reads from raw data. Reference genome and gene model annotation files were downloaded from genome website directly. Index of the reference genome was built using STAR and paired-end clean reads were aligned to the reference genome using STAR (v2.5.1b). STAR uses the method of Maximal Mappable Prefix (MMP). HTSeq v0.6.0 was used to count the read numbers mapped to each gene.
Analysis of differential expression was performed using the edgeR package (3.12.1). The resulting P values were adjusted using the Benjamini–Hochberg method for controlling the false discovery rate. Genes with an adjusted P value < 0.05 found by DESeq2 were assigned as differentially expressed. The hierarchical clustering heat map was generated with the ggplot library. Gene Ontology (GO) as well as Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis of differentially expressed genes was implemented by the cluster Profiler R package, in which gene length bias was corrected. GO terms and KEGG pathways with corrected P value < 0.05 were considered significantly enriched in differential expressed genes.
Mitochondrial respiration in permeabilized myocardial fibers
Approximately 10 mg intact myocardial tissue adjacent to border zone was dissected from each heart. The cardiac tissues were transferred into a petri dish with ice-cold BIOPS [35]. The tissues were separated mechanically into fiber bundles with two pairs of very sharp forceps, observing a change from red to pale coloring of the separated fiber bundles. Then the fiber bundles were permeabilized by gentle agitation for 30 min at 4 °C in ice cold BIOPS supplemented with 50 mg/ml saponin [61]. Myofibers were washed for 10 min by agitation in ice-cold mitochondrial respiration medium MiR05, blotted, weighed (Mettler Toledo microbalance XS205DU), and immediately used for respirometric measurements. Respiration was measured at 37 °C, with 2 mg permeabilized fibers in each high-resolution respirometry chamber containing 2 mL MiR05, using 2 chambers in parallel (OROBOROS Oxygraph-2k, Innsbruck, Austria). DatLab software (OROBOROS INSTRUMENTS) was used for data acquisition and analysis.
ATP content detection
Myocardial ATP content detection was performed on fresh, infarcted heart tissues according to the manual of the Enhanced ATP Assay Kit (S0027; Beyonetime). Briefly, ATP was extracted by lysis buffer and measured by a luciferase reaction in which 560 nm light was emitted when D-luciferin was converted to oxyluciferin. Luminescence was measured using a luminometer (SpectraMax M5/M5e; Molecular Devices). ATP level was calculated according to a calibration curve with standard ATP samples. The total amount of tissue protein was used for normalization to ATP contents.
Quantification of purine nucleotides
Myocardial purine nucleotides were extracted from frozen tissue samples with 0.6 M perchloric acid [1]. The extraction mixture was centrifuged for 10 min at 10,000 g and 4 °C, the supernatant was taken and quickly neutralized to pH = 6.5 ~ 7 with 1 M KOH solution. The neutralized supernatant was then allowed to stand for 5 min in an ice bath to precipitate the potassium perchlorate and then centrifuged for 10 min at 10,000 g and 4 °C. The neutralized supernatant was filtered through a 0.22-µm filter.
Purine nucleotides from the infarcted hearts were detected by HPLC (LC-20; SHIMADU) [18]. The purine nucleosides were separated on Ultimate AQ-C18 columns (250 × 4.6 mm). Peaks were detected and analyzed at 254 nm by a SPD-M20A diode array detector. Mobile phase A consisted of 60 mM dipotassium hydrogen phosphate and 40 mM potassium dihydrogen phosphate dissolved in deionized water, pH = 7, while mobile phase B consisted of 100% acetonitrile. HPLC separation was achieved using continuous gradient elution: 0 min 100% A, 0% B; 2 min 95% A, 5% B; 4 min 80% A, 20% B; 5.3 min 75% A, 25% B and 6 min 100% A, 0% B. Flow rate of the mobile phase was 1.2 mL/min, the injection volume was 20 µL. ATP, ADP and AMP in the samples were identified by comparison with retention time of standards, while the concentrations of ATP, ADP and AMP were determined according to the standard curve. The protein was dissolved in 50 mM NaOH at 4 °C overnight, and protein concentration was measured by BCA methods.
Tagged-RNA pulldown
To identify the binding partners of LncHrt, we used a tagged-RNA pulldown assay, as described [37, 64]. Sense and antisense strands of streptavidin-binding S1m DNA were synthesized (Tsingke), annealed, and cloned into pCDH-MSCV at a cloning site, which was then used to insert sequences encoding LncHrt. The constructs expressing S1m, S1m-LncHrt as well as those expressing untagged LncHrt and EGFP were packaged into lentivirus using PEI (1:4) and then infected the cardiac fibroblasts with 20 MOI for 48 h. Cells were then harvested in SA-RNP lysis buffer (20 mM Tris–HCl (pH 7.5), 150 mM NaCl, 1.5 mM MgCl2, 2 mM DTT, 50 U/mL RNase OUT (Life Technologies) 50 U/mL Superase IN (Ambion) and 1 × complete protease inhibitor tablet (Roche). Streptavidin–Sepharose beads were blocked with 500 ng/µL yeast tRNA and 1 mg/mL BSA in SA-RNP lysis buffer before being added into cell lysates and being incubated at 37 °C for 2 h on a rotator. The beads were then pelleted and washed five times with SA-RNP washing buffer (20 mM Tris–HCl (pH 7.5), 300 mM NaCl, 5 mM MgCl2, 2 mM DTT, 50 U/mL RNase OUT (Life Technologies, NY, USA), 50 U/mL Superase IN (Ambion) and 1 × complete protease inhibitor tablet (Roche). After the last wash, RNA-bound proteins were then boiled in 50 µL 3 × LDS sample buffer (Life Technologies) and used for mass spectrometry.
RNA immunoprecipitation (RIP)
RIP experiments were performed with the Magna RIPTM RNA-Binding Protein Immunoprecipitation Kit (Merck Millipore) as directed by the manufacturer. Briefly, 1-month-old mouse heart tissues were lysed thoroughly on ice in RIP Lysis Buffer supplemented with protease inhibitor cocktail and RNase inhibitor using a glass homogenizer to facilitate lysis. CDK5 and SIRT2 antibodies (Supplementary Table 8) with corresponding IgG were incubated with magnetic beads at room temperature for 1 h for each immunoprecipitation. After a 13,000 g centrifugation for 10 min, supernatants were incubated with the indicated antibody-bead complexes overnight at 4 °C with rotation. Co-immunoprecipitated RNAs were extracted as described by the kit manual, reverse-transcribed to cDNA, then subjected to qRT-PCR to measure LncHrt enrichment using primers listed in Supplementary Table 4. IgG enrichment served as negative controls.
Immunoprecipitation (IP) and co-immunoprecipitation (Co-IP)
Heart tissues were lysed with immunoprecipitation (IP) buffer (Thermo) supplied with protease inhibitor cocktail (Roche) at 4 °C using a glass homogenizer to facilitate tissue lysis. Samples were then centrifuged at 13,000 g for 20 min, the lysates were precleared and 3 mg total protein/IP was incubated with the indicated primary antibodies overnight at 4 °C with rotation before incubation with Protein G Dynabeads (Invitrogen) for 6 h at 4 °C. The beads were then washed with high salt buffer once and IP buffer five times. 100 µL 3 × SDS loading buffer was added to the beads and boiled at 95 °C for 10 min prior to Western blot detection.
Western blot analysis
Protein lysates were prepared from heart tissues in tissue extraction reagent (Invitrogen) supplemented with proteinase inhibitors. Cultured cells were harvested, homogenized, and incubated in Cell Extraction Buffer (Invitrogen) with protease inhibitor cocktail (Sigma) and 1 mM phenylmethyl sulfonyl fluoride (PMSF). Samples (50 µg total protein for each) were separated by 10% SDS–PAGE and electrophoretically transferred onto PVDF membranes. After blocking in 5% skimmed milk, membranes were incubated overnight at 4 °C with the indicated primary antibodies and then washed three times with TBST buffer before incubation for 1 h with HRP-conjugated secondary antibodies at room temperature. Protein bands were visualized using ECL Reagent (Invitrogen) with the Bio-Rad ChemiDoc imaging system. All the antibody information is listed in Supplementary Table 14.
Statistical analysis
Statistical significance between two columns was assessed by two-tailed Student’s t test or Mann–Whitney test; for more than two columns, one-way ANOVA (Dunnett’s multiple comparisons test) or two-way ANOVA (Bonferroni's multiple comparisons test) analysis was used. Unless otherwise stated, results are presented as mean value ± standard error of the means (SEM).
Results
LncHrt is a cardiomyocyte-enriched lncRNA downregulated following myocardial infarction
To explore pivotal lncRNAs that regulate pathophysiological processes in heart disease, we performed microarray-based transcriptome profiling on mouse hearts 3 and 14 days after myocardial infarction (MI) and compared these to sham-operated control mice. An earlier report showed that Cfast is one of the most upregulated lncRNAs in MI hearts, and it regulates cardiac fibrotic remodeling following cardiac injury [73].
The present study focused on downregulated lncRNAs. Transcriptome analysis revealed 543 lncRNAs downregulated (fold change > 2, P < 0.05) in mouse hearts 3 days post-MI (MI-3d, Fig. 1a), and 62 lncRNAs downregulated (fold change > 2, P < 0.05) in hearts 14 days post-MI (MI-14d, Fig. 1b). Using combined analyses, we identified a total of 36 lncRNAs downregulated in both MI-3d and MI-14d time points (Fig. 1c, d and Supplementary Table1). Among them, the top five most downregulated lncRNAs are AK034241, NR_045336, XR_373578, NR_131053, and XR_386048. In particular, the expression of AK034241 decreased over 22-fold (Fig. 1a–d and Supplementary Table 1).
LncHrt is a cardiomyocyte-enriched lncRNA downregulated after myocardial infarction. a Volcano plot of downregulated lncRNAs in the heart tissues 3 days post-MI (MI-3d), Fold Change > 2, P value < 0.05. b Volcano plot of lncRNAs downregulated in the heart tissues 14 days post-MI (MI-14d), Fold Change > 2, P value < 0.05. c Venn plot of down regulated lncRNAs between MI-3d and MI-14d. d Heatmap of 36 downregulated lncRNAs in both MI-3d and MI-14d. e qRT-PCR of gene expression of 5 top downregulated lncRNAs (AK034241, NR045336, XR373578, NR0131053, XR386048) in the heart 3 days and 14 days after MI. Rn18S as a reference gene. Data are mean ± s.e.m. **P < 0.01 and ***P < 0.001 (versus Sham); by One-way ANOVA analysis, n = 6 mice per group. f Gene expression of 5 top downregulated lncRNAs in different mouse tissues by qRT-PCR. Rn18S as a reference gene. Data are mean ± s.e.m. *P < 0.05, **P < 0.01 and ***P < 0.001 (versus Heart); by One-way ANOVA analysis, n = 3 mice per group. g Gene expression of 5 top downregulated lncRNAs in heart tissues of postnatal 21 days (P21d), 1 month (1 M) and 2 months (2 M) old mice. Rn18S as a reference gene. Data are mean ± s.e.m. *P < 0.05, **P < 0.01 and ***P < 0.001 (versus LncHrt); by One-way ANOVA analysis, n = 4 mice per group. h Image of agarose electrophoresis showing RT-PCR of AK034241, NR045336, XR373578, NR0131053, XR386048 in P21d, 1 M and 2 M old heart tissues. i LncHrt expression in cardiomyocytes and non-cardiomyocytes. Rn18S as a reference gene. Data are mean ± s.e.m. *P < 0.05 and **P < 0.01 (versus Cardiomyocytes); by Student’s t test, n = 6 mice per group. j Subcellular location of LncHrt in cardiomyocytes. Actb acts as a cytoplasm marker while U6 as a nuclear marker. k Coding probability of LncHrt predicted by Coding Potential Calculator (CPC). Protein coding mRNAs Cdk1 and Gapdh serve as negative controls and reported LncRNAs Chaer and Hotair are positive controls. l Conservation analysis of LncHrt in different mammalian species in UCSC Genome Browser on Mouse Dec. 2011 (GRCm38/mm10) Assembly. m qRT-PCR detection of LncHrt gene expression in mice TAC model. Rn18S as a reference gene. n = 3 or 5 per group. n qRT-PCR detection of the human homology LncHrt gene expression in human hearts with dilated cardiomyopathy (DCM). Rn18S as a reference gene. n = 2 or 6 human heart samples per group
We validated these top five downregulated lncRNAs using an independent batch of MI heart tissues by quantitative reverse transcription polymerase chain reaction (qRT-PCR) (Fig. 1e). To determine the tissue distribution of these lncRNAs, we assessed their expression in different tissues. We found that AK034241 and XR_373578 was specifically enriched in the heart (Fig. 1f), indicating their potential relevance in cardiac disease. Intriguingly, AK034241 was the most abundantly expressed lncRNA in heart tissue compared to the other 4 lncRNAs (Fig. 1g, h). As a result, we focused on AK034241 and named it lncRNA-heart (LncHrt).
Next, we investigated the developmental expression pattern of LncHrt (i.e., from neonates to adulthood) and found that LncHrt was progressively elevated 7 days after birth and then dramatically increased until adulthood, indicating LncHrt may be essential for adult heart homeostasis. (Fig. S1, a, b). To determine the cell type expressing LncHrt, we isolated cardiomyocytes and non-cardiomyocytes from adult mouse hearts and found LncHrt was primarily expressed in adult cardiomyocytes (Fig. 1i). In addition, we performed fractionation assays to determine the subcellular location of LncHrt. qRT-PCR assays detected that LncHrt was equally distributed in the nucleus and cytoplasm of cardiomyocytes (Fig. 1j), suggesting that LncHrt may participate in both transcriptional and post-transcriptional regulatory processes.
We performed 5ʹ and 3ʹ rapid amplification of cDNA ends (RACE) using RNA isolated from adult mouse hearts and demonstrated the full length of LncHrt is 1907 nt (Fig. S1, c, d). There were an additional 9 nt in the 5ʹ end compared to annotated AK034241. The LncHrt gene contains two exons, and like many lncRNAs, it has a poly(A) tail. We evaluated if the LncHrt transcript is a true non-coding RNA using the Coding Potential Calculator [30] and Coding Potential Assessment Tool [63]. As expected, both revealed that the LncHrt transcript has a very low coding-potential score, similar to Chaer [64] and Hotair [13], two well-known lncRNAs, and unlike protein-coding genes, like Cdk1 and Gapdh, which have high coding-potential scores (Fig. 1k and Fig. S1e).
We subsequently examined the conservation of LncHrt using the UCSC Genome Browser (GRCm38/mm10). The genome locus of LncHrt in humans is conserved, while the LncHrt sequence is highly conserved across multiple species in exon 1 as well as the 5ʹ-end and 3ʹ-end of exon 2 (Fig. 1l). The predicted LncHrt secondary structure based on minimum free energy (MFE) and partition function was assessed using the RegRNA Server (http://rna.tbi.univie.ac.at/cgi-bin/RNAWebSuite/RNAfold.cgi). These analyses revealed that LncHrt has stem-loop structures with relatively high base-pairing potential (Figure. S1f).
Consistent with the finding that mouse LncHrt was downregulated in the heart after myocardial infarction, we also found that LncHrt was decreased in the murine transverse aortic constriction (TAC) model and the expression of the human homolog of mouse LncHrt was reduced in patients with dilated cardiomyopathy (DCM) (Fig. 1m, n and Supplementary Table 2). These findings suggest that this lncRNA may play an important role in the regulation of heart disease.
Knock-down of LncHrt impairs cardiac homeostasis
To define the in vivo function of LncHrt in the heart, we knocked down (KD) LncHrt expression using AAV9-shRNA (short hairpin RNA) directed against LncHrt (AAV-LncHrt KD) in neonatal mice, as previously reported [10]. For the control group, the mice received an AAV9 expressing a scrambled short hairpin RNA sequence (AAV-Scramble) (Fig. 2a). AAV-LncHrt KD resulted in a decreased cardiac expression of LncHrt by 75% compared to AAV-Scramble (Fig. 2b) and the hearts of AAV-LncHrt KD mice were enlarged compared to control mice (Fig. 2c). The heart weight to body weight ratio (HW/BW) was also significantly increased in these mice (Fig. 2d) with no effect on mice body weight (Fig. 2e).
Knock-down of LncHrt impairs cardiac homeostasis. a Schematic diagram showing the experimental procedures for AAV9 delivered LncHrt knocking down (KD) in mice. b qRT-PCR of LncHrt gene expression. Rn18S as a reference gene. Data are mean ± s.e.m. *P < 0.05 and **P < 0.01 (versus AAV-Scramble); by Mann–Whitney test, n = 6 mice per group. c Representative images of gross morphology of hearts from AAV-Scramble and AAV-LncHrt KD. Scale bar = 2 mm. d Ratio of heart weight (HW) to body weight (BW) in AAV-Scramble and AAV-LncHrt KD heart at 2–3 months. Data are mean ± s.e.m. *P < 0.05 and **P < 0.01 (versus AAV-Scramble); by Student’s t test, n = 6 mouse per group. e Body weight (BW) in AAV-Scramble and AAV-LncHrt KD heart at 2–3 months. Data are mean ± s.e.m. *P < 0.05 and **P < 0.01 (versus AAV-Scramble); by Student’s t test, n = 6 mice per group. f M-mode echocardiography of mice heart 2–3 months after AAV9 delivered LncHrt-shRNA injection. g-i Echocardiography analyses of cardiac function of FS% (g), LVID (h) and. LVPW (i) after AAV9 delivered LncHrt-shRNA 2–3 months compared to their scramble group. FS, left ventricular fractional shortening. LVID,s, LV internal dimension at end-systole. LVPWs, left ventricular posterior wall at end-systole. Data are mean ± s.e.m. *P < 0.05 and **P < 0.01 (AAV-Scramble); by Student’s t test, n = 7 mice per group. j Representative of low- (top) and high-magnification (bottom) of images of Sirius red & fast green staining of scramble and LncHrt knock-down heart sections, Scale bars, 2 mm (up), 50 μM (down). k Quantification of collagen volume of scramble and LncHrt knocking down hearts. Data are mean ± s.e.m. *P < 0.05 and **P < 0.01 (versus AAV-Scramble); by Student’s t test, n = 4 mice. l Representative pictures of cardiomyocytes stained with WGA. Scale bars = 100 µm. m Quantification of cardiomyocyte area of AAV-LncHrt KD compared to AAV-Scramble group. Data are mean ± s.e.m. ***P < 0.001 (versus AAV-Scramble); by Student’s t test, n = 5 mice hearts per group, measured ~ 400 cardiomyocytes per heart and calculated median value for each heart. n Gene expression of cardiac disease markers in scramble and LncHrt knock down hearts. Rn18S as a reference gene. Data are mean ± s.e.m. *P < 0.05 and **P < 0.01 (versus AAV-Scramble); by Student’s t test (Nppa) or Mann–Whitney test (Nppb, Myh7/Myh6), n = 6 mice per group
Echocardiography revealed that AAV-LncHrt KD impaired left ventricular systolic heart function (Fig. 2f and Supplementary Table 3) and decreased fractional shortening (Fig. 2g). In particular, AAV-LncHrt KD hearts exhibited a dilated left ventricular chamber [LVID,s (mm), Fig. 2h and Supplementary Table 3] and a reduced left ventricular posterior wall thickness [LVPW,s (mm), Fig. 2i and Supplementary Table 3].
We validated these structural changes histologically and assessed fibrosis in AAV-Scramble and AAV-LncHrt KD treated hearts (Fig. 2j, k). These hearts were also used to assess cardiomyocyte size (Fig. 2l and m). Marker analyses demonstrated a significant increase in the expression of cardiomyopathy marker genes such as Nppa, Nppb, and Mhy7/Myh6 after LncHrt knockdown (Fig. 2n), which supports our observation that knock-down of LncHrt adversely affects cardiac function. To confirm this, and rule out the possibility of off-target effects resulting from shRNA-mediated LncHrt knock-down, we designed another LncHrt shRNA to target a different site of the LncHrt gene (AAV-LncHrt KD-A) and obtained similar results (i.e., impaired heart function and increased pathological remodeling) (Fig. S2, a–n and Supplementary Table 3). Together, these data indicate that LncHrt plays an essential role in maintaining normal cardiac function.
LncHrt overexpression protects the heart from myocardial infarction
Because inhibition of LncHrt expression negatively impacts cardiac function, and decreased LncHrt levels were found in the hearts after MI, we tested the hypothesis that overexpression of LncHrt could play a protective role in the pathological progression of MI. We employed AAV9-mediated delivery system to overexpress LncHrt in vivo using the cTNT promoter to achieve cardiomyocyte-specific overexpression of LncHrt (AAV-LncHrt). An AAV9-luciferase construct was used as a control (AAV-CTRL) (Fig. 3a). qRT-PCR analysis confirmed increased LncHrt cardiac expression 2 months after AAV-LncHrt treatment (Fig. 3b). Triphenyltetrazolium chloride (TTC) viability staining validated that MI induced infarct size is comparable between the two groups (Fig. S3, a, b). Overexpression of LncHrt improved cardiac morphology and significantly reduced the HW/BW ratio in response to MI compared to the control group (Fig. 3c, d) without affecting mice body weight (Fig. 3e). More importantly, overexpression of LncHrt ameliorated cardiac function after MI (Fig. 3f), as determined by echocardiography that showed improved fractional shortening (FS%) (Fig. 3g and Supplementary Table 4), maintenance of left ventricular posterior wall thickness [LVPW,s (mm), Fig. 3h and Supplementary Table 4], and reduced cardiac chamber internal diameter [LVID,s (mm), Fig. 3i and Supplementary Table 4].
AAV9-delivered LncHrt overexpression protects the heart from myocardial infarction. a Schematic diagram showing the experimental design with AAV9-delivered LncHrt overexpression in mice hearts with myocardial infarction injury. Echocardiography was performed at Pre-MI, post-MI 1 month (MI-1M) and 2 months (MI-2M). b qRT-PCR of LncHrt gene expression after MI-2M. Rn18S as a reference gene. Data are mean ± s.e.m. *P < 0.05, **P < 0.01, ***P < 0.001 (versus AAV-CTRL); by Mann–Whitney test, n = 14–16 mice per group. c Representative images of gross morphology of hearts from AAV-CTRL and AAV-LncHrt. Scale bar = 2 mm. d Ratio of heart weight (HW) to body weight (BW) in AAV-CTRL and AAV-LncHrt heart post-MI (2 months). Data are mean ± s.e.m. *P < 0.05 and **P < 0.01 (versus AAV-CTRL); by Student’s t test, n = 10–11 mouse per group. e Body weight (BW) in AAV-CTRL and AAV-LncHrt heart post-MI (2 months). Data are mean ± s.e.m. *P < 0.05 and **P < 0.01 (versus AAV-CTRL); by Student’s t test, n = 10–11 mice per group. f M-mode echocardiography of mice heart 2 months after AAV9 delivered LncHrt injection. g-i Analyses of cardiac function of FS% (g), LVPW (h) and LVID (i) after AAV-LncHrt injection at pre-MI, 1 and 2 months post-MI compared to their CTRL group. FS, left ventricular fractional shortening. LVPWs, left ventricular posterior wall at end-systole. LVIDs, LV internal dimension at end-systole. Data are mean ± s.e.m. *P < 0.05 and **P < 0.01 (versus AAV-CTRL); by 2-way ANOVA analysis, n = 14–16 mice per group. j Representative of images of Sirius red/fast green collagen staining of series of transverse sections after injection of AAV-LncHrt compared to AAV-CTRL group after MI injury. Sirius red/fast green collagen staining marks myocardium (green) and scar (red). Scale bars = 2 mm. k Quantification of scar size of AAV-LncHrt compared to AAV-CTRL group after MI injury. Data are mean ± s.e.m. *P < 0.05 and **P < 0.01 (versus AAV-CTRL); by Student’s t test, n = 7 mice per group. l Representative images of cardiomyocytes stained with WGA. Scale bars = 100 μm. m Quantification of cardiomyocytes area of AAV-LncHrt compared to AAV-CTRL group after MI injury. Data are mean ± s.e.m. *P < 0.05, **P < 0.01 and ***P < 0.001 (versus AAV-CTRL); by Student’s t test, n = 5 mouse hearts per group, measured ~ 300 cardiomyocytes per heart and calculated median value for each heart. n Hypertrophic marker gene expression of heart of AAV-LncHrt versus AAV-CTRL group. Rn18S as a reference gene. Data are mean ± s.e.m. *P < 0.05 and **P < 0.01 (versus AAV-CTRL); by Student’s t test or Mann–Whitney test, n = 7 mice per group
Histological analyses revealed that overexpression of LncHrt in the heart substantially reduced infarct fibrosis following MI when compared to controls (Fig. 3j, k). In addition, cardiomyocyte size was markedly decreased in AAV-LncHrt treated hearts post-MI (Fig. 3l, m), indicating reduced cardiac pathological remodeling. Molecular marker analyses demonstrated a significant decrease in the expression of the hypertrophic and cardiomyopathy marker genes Nppa, Nppb, and Myh7/Myh6 in AAV-LncHrt-treated hearts (Fig. 3n). We also assessed the effects of LncHrt on healthy mouse hearts as a baseline. We found that LncHrt did not affect cardiac function and heart morphology in healthy hearts (Fig. S4, a–j and Supplementary Table 5). Together, these results indicate that LncHrt plays a role in preserving cardiac function and attenuates adverse remodeling in response to MI.
LncHrt overexpression rescues the transcriptome in infarcted hearts
To gain a full understanding of the molecular mechanisms underlying LncHrt-mediated cardiac protection following MI, we performed genome-wide RNA sequencing (RNA-seq) on ventricular tissue from AAV9-mediated LncHrt overexpression (AAV-LncHrt) and control (AAV-CTRL) hearts 7 days post-MI (Fig. 4a). Boxplot, hierarchical clustering of the RNA-sequencing data, and PCA analysis confirmed consistency between the biological repeats for each group (n = 3 for each experimental group. Fig. S5, a–c). Hierarchical clustering analyses indicated that overexpression of LncHrt markedly altered the cardiac transcriptome following MI (Fig. 4b).
Cardiac LncHrt overexpression rescued transcriptomes of myocardial infarction. a Schematic diagram showing the experimental design of RNA-sequencing (seq) in AAV-LncHrt vs. CTRL post-MI, n = 3 mice per group. b Hierarchical clustering of differentially expressed genes in AAV-LncHrt vs. CTRL hearts as assessed by RNA-seq (adjusted P value < 0.05). c Volcano plot of differentially expressed genes in AAV-LncHrt vs. CTRL hearts (Log2Fold Change > 0.55, adjusted P Value < 0.05). d KEGG pathway enrichment analysis of differentially expressed genes in AAV-LncHrt vs. CTRL hearts post myocardial infarction using DAVID tools. e Gene Ontology (GO) analysis linked to biological process of differentially expressed genes in AAV-LncHrt vs. CTRL hearts post myocardial infarction using DAVID tools. f Schematic diagram showing the experimental design of combined analysis of AAV-LncHrt RNA-seq post-MI and Micro-array of MI heart by Gene set enrichment analysis (GSEA). g GSEA analysis of metabolic related enrichment plots which inhibited in MI were rescued by LncHrt. h Heat maps of metabolic related genes validation by qRT-PCR using an independent batch of samples post-MI-7d and MI-2 M
We identified 1763 differentially expressed genes (DEGs, adjust P value < 0.05) between the LncHrt overexpression and control group subjected to MI, in which 1083 genes were upregulated and 680 genes were downregulated (Fig. 4c). We then analyzed the functional annotation of these differentially expressed genes using Database for Annotation and Visualization and Integrated Discovery (DAVID) tools. Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses revealed upregulated genes are largely related to metabolic-related pathways, namely “metabolic pathways”, “carbon metabolism”, “oxidative phosphorylation”, “the citric acid (TCA) cycle”, “glycolysis”, and “fatty acid metabolism”. The “cardiac muscle contraction” related pathway was also upregulated. In contrast, downregulated genes are related to cardiac pathological pathways, such as “ECM-receptor interaction” and “focal adhesion” (Fig. 4d). Similarly, Gene Ontology (GO) biological process analyses confirmed that “metabolic processes” and “cardiac muscle contraction” are enriched among upregulated genes while “immune system process”, “inflammatory response”, and “collagen fibril organization” are enriched in downregulated genes (Fig. 4e).
These findings indicate that LncHrt overexpression-improved heart function post-Ml likely, by mediating cardiac metabolic pathways. To test this hypothesis, we performed comparative analyses of transcriptome signatures from both control and MI hearts with or without LncHrt overexpression (Fig. 4f). Gene Set Enrichment Analysis (GSEA) demonstrated that MI-induced down-regulation of extensive metabolic processes such as “oxidative phosphorylation”, “fatty acid metabolism”, and “glycolysis” were completely restored upon LncHrt overexpression compared to control post-MI (Fig. 4g). Cardiac pathological pathways like “ECM receptor interaction” and “TGF beta signaling pathway” were repressed by LncHrt in MI hearts and “cardiac muscle contraction” was increased (Fig. 4f and Fig. S5d). These analyses support the notion that LncHrt plays an important role in cardiac metabolism and muscle contraction following MI.
To confirm these results experimentally, we collected heart samples from an independent set of animals and performed qRT-PCR analysis for differentially expressed signature genes. Our results validated the changes described above in multiple pathways including “carbon metabolism”, “TCA cycle”, “fatty acid metabolism”, “glycolysis”, “fatty acid degradation”, “pyruvate metabolism”, and “cardiac muscle contraction” (Fig. 4h). In contrast, knock-down of LncHrt resulted in an opposite effect on metabolic gene expression (Fig. S5e). Together, these data indicate LncHrt positively modulates metabolic-related gene expression, enhanced cardiac muscle contraction, and suppressed cardiac fibrosis, which may improve cardiac function post-MI.
LncHrt improved cardiac metabolic homeostasis post-MI
The metabolic processes modulated by LncHrt post-MI identified by RNA-seq analysis prompted us to investigate how LncHrt mitigates cardiac metabolic stress and improve metabolic phenotype in response to cardiac remodeling and stress (Fig. 5a). Cardiac LncHrt overexpression was confirmed by RT-PCR in mouse hearts after AAV-LncHrt injection (Fig. 5b). We assessed mitochondrial respiration by measuring oxygen consumption rate (OCR) in permeabilized mouse myofibers in the remote ischemic zone using high-resolution respirometry following substrate-uncoupler-inhibitor-titration (SUIT) protocols [34, 41]. ADP-stimulated oxygen flux was determined under coupled conditions to estimate mitochondrial function, while oxygen flux was enhanced in LncHrt-injected hearts (Fig. 5c and Fig. S6a). Respiratory stimulation of the fatty acid oxidation (FAO) pathway in the presence of malate (M) and octanoylcarnitine (Oct) was significantly elevated by LncHrt (Fig. 5d). Combined analyses of the oxidative phosphorylation (OXPHOS) capacity of mitochondria in an electron transfer (ET) state, with or without fatty acid oxidation (F-pathway) were performed (Fig. 5c and Fig. S6a). LncHrt overexpression significantly promoted complex I OXPHOS capacity in the presence of pyruvate (P), glutamate (G), and malate (M) to 165%, and the complex II oxygen consumption rate (OCR) increased to 154%, when compared to the CTRL group, respectively (Fig. 5e). By adding ascorbate (As) and TMPD, the respiration of complex IV was increased to 131% in LncHrt-overexpressing hearts, when compared with controls (Fig. 5f). We also assessed mitochondrial content by examining the protein level of the mitochondrial proteins, VDAC and COXIV. We found that LncHrt overexpression did not alter the mitochondrial content, indicating the beneficial effect of LncHrt on mitochondrial respiration were not from a higher content of mitochondria in the preparations used for respiration measurement (Fig. S6, b, c). Together, these studies indicate cardiac overexpression of LncHrt improves oxidative phosphorylation metabolic capacity in the heart after MI.
LncHrt improved cardiac metabolic homeostasis post myocardial infarction. a Schematic plot showing the experimental design with AAV9 delivered LncHrt overexpression in mice heart with myocardial infarction injury, mitochondrial respiration capacity and glucose utilization was performed from 1 to 2 weeks post-MI (MI-1 W and MI-2 W). b qRT-PCR of LncHrt gene expression between MI-1 W and MI-2 W. Rn18S as a reference gene. Data are mean ± s.e.m. *P < 0.05 and **P < 0.01 (versus AAV-CTRL); by Mann–Whitney test, n = 11 mice per group. c Representative respiratory experiment of mitochondrial oxidative phosphorylation (OXPHOS) capacity of complex with fatty acid oxidation pathway(F-pathway) in OXPHOS state by using application of substrate uncoupler inhibitor titration (SUIT) protocols to interrogate sequentially different substrate and coupling states using saponin permeabilized myofibers. d Fatty acid oxidation (FAO) as measured by oxygen consumption with octanoylcarnitine corrected to corresponding residual oxygen consumption (ROX). Data are mean ± s.e.m. *P < 0.05 and **P < 0.01 (versus AAV-CTRL); by Student’s t test, n = 7 mice per group. e Combined analysis of mitochondrial oxidative phosphorylation (OXPHOS) capacity of complex I (CI) and complex II (CII) in electron transfer (ET) state with or without fatty acid oxidation pathway(F-pathway) in OXPHOS state. The oxygen consumption rate (OCR) of complex I showed oxygen consumption with Pyruvate (P), Malate (M) and Glutamate (G) (PMG) titrations minus octanoylcarnitine (OCT) titrations or corrected to corresponding residual oxygen consumption (ROX), while complex II respiration rate corrected to corresponding ROX. Data are mean ± s.e.m. *P < 0.05 and **P < 0.01 (versus AAV-CTRL); by Student’s t test, n = 11 mice per group. f Combined analysis of mitochondrial oxidative phosphorylation (OXPHOS) capacity of complex IV (CIV) in electron transfer (ET) state with or without fatty acid oxidation pathway(F-pathway) in OXPHOS state. As illustrated by oxygen consumption rate (OCR) corrected to corresponding residual oxygen consumption (ROX). Data are mean ± s.e.m. *P < 0.05 and **P < 0.01 (versus AAV-CTRL); by Student’s t test, n = 11 mice per group. g Western blot of key enzymes of glycolysis and TCA cycle in infarcted myocardium. h Quantification of Western blot band density using image J. Data are mean ± s.e.m. *P < 0.05 and **P < 0.01 (versus AAV-CTRL); by Student’s t test, n = 4 mice per group. i ATP content in infarct heart tissues normalize to protein levels. Data are mean ± s.e.m. *P < 0.05 and **P < 0.01 (versus AAV-CTRL); by Student’s t test, n = 9–10 mice per group. j Representative respiratory experiment of mitochondrial oxidative phosphorylation (OXPHOS) capacity of complex with fatty acid oxidation pathway(F-pathway) in OXPHOS state by using application of substrate uncoupler inhibitor titration (SUIT) protocols to interrogate sequentially different substrate and coupling states using saponin permeabilized myofibers (AAV-LncHrt KD versus AAV-Scramble). k Fatty acid oxidation (FAO), as measured by oxygen consumption with octanoylcarnitine corrected to corresponding residual oxygen consumption (ROX). Data are mean ± s.e.m. *P < 0.05 and **P < 0.01 (versus AAV-Scramble); by Student’s t test, n = 4 mice per group. l Mitochondrial respiration of complex I (CI) and complex II (CII). The oxygen consumption rate (OCR) of complex I showed oxygen consumption with Pyruvate (P), Malate (M) and Glutamate (G) (PMG) titrations minus octanoylcarnitine titrations, while complex II respiration rate corrected to corresponding residual oxygen consumption (ROX). Data are mean ± s.e.m. *P < 0.05 and **P < 0.01 (versus AAV-Scramble); by Student’s t test, n = 4 mice per group. m Mitochondrial oxidative phosphorylation (OXPHOS) capacity of complex IV (CIV). As illustrated by oxygen consumption rate (OCR) corrected to corresponding residual oxygen consumption (ROX). Data are mean ± s.e.m. *P < 0.05 and **P < 0.01 (versus AAV-Scramble); by Student’s t test, n = 4 mice per group
Given that reductions in the supply of oxygen following MI elicits a metabolic shift to glucose utilization [29], we examined the protein levels of hexokinase-1 (HK1) [16] and phosphoglycerate kinase 1 (PGK1) [38], which are both key glycolytic enzymes, as well as citrate synthase (CS) [8, 23], isocitrate dehydrogenase (IDH2) [31], and energy production enzymes in the infarct of hearts under hypoxic conditions. We found a marked increase in the expression of these proteins in LncHrt overexpressing hearts (Fig. 5g, h). These results indicate that LncHrt overexpression protected mitochondrial oxidative metabolism after MI.
Next, we determined if LncHrt increased cardiac ATP content after MI. LncHrt overexpression markedly enhanced ATP levels by 160% compared to CTRL hearts after MI (Fig. 5i). Conversely, we asked whether knock-down of LncHrt also impacts metabolic homeostasis in the heart. We measured key metabolic pathways involved in cardiac contraction using permeabilized mouse myofibers. As expected, we observed impaired mitochondrial respiration (Fig. 5j), fatty acid oxidation (Fig. 5k), and complex I oxidative phosphorylation capacity in the LncHrt knockdown group compared to the control group with mild inhibition of complex II and complex IV oxidative phosphorylation capacity (Fig. 5l, m). Together, our results show LncHrt preserved cardiac metabolic homeostasis and improved cardiac function after MI.
LncHrt activates the LKB1-AMPK signaling pathway via SIRT2
To elucidate the molecular mechanism by which LncHrt modulates myocardial metabolic homeostasis in response to MI, we first asked whether LncHrt may regulate the expression of its neighboring gene, Klhl33. We overexpressed LncHrt to assess the transcriptional impact on Klhl33 in vivo and in vitro. qRT-PCR analysis showed that LncHrt overexpression had no effect on Klhl33 gene expression in vivo before (Fig.S7a) or after MI (Fig.S7b) and overexpression of LncHrt did not affect Klhl33 levels in vitro (Fig.S7c).
We identified LncHrt-interacting proteins by fusing a modified streptavidin (SA)-binding S1 RNA tag, termed S1m [37], to full-length LncHrt and expressing this construct in cardiac fibroblasts. The S1m construct lacking LncHrt served as a negative control. Then, streptavidin-conjugated magnetic beads were used in an RNA pull-down assay, and putative LncHrt-interacting proteins were identified by mass spectrometry (Fig. 6a). From this screening, we found many putative LncHrt-interacting proteins. At the top of this list is NAD-dependent deacetylase sirtuin 2, encoded by the Sirt2 gene (Fig. 6b and Supplementary Table 6). We investigated SIRT2 because it is a crucial metabolic modulator that acts as a cardioprotective deacetylase in pathological hypertrophy [53, 58, 72]. We validated that LncHrt interacted with SIRT2 in cardiomyocytes using a S1m-fused LncHrt pull-down assay (Fig. 6c). To confirm the interaction between LncHrt and SIRT2 endogenously, we performed an RNA immunoprecipitation (RIP) assay. Anti-SIRT2 antibody was incubated with heart tissue lysates and co-precipitated LncHrt was analyzed by qRT-PCR, using two primer sets to detect two independent exons of the LncHrt gene. We observed enrichment of LncHrt in SIRT2 immunoprecipitate compared to IgG control (Fig. 6d, e), indicating an association between LncHrt and SIRT2 in the heart.
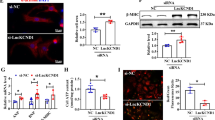
LncHrt activates the LKB1-AMPK signaling pathway by interacting with SIRT2. a Workflow of S1m tagged LncHrt pull down assay in cardiac cells. b Amino acid of the protein SIRT2. The detected peptide by mass spectrometry is highlighted in yellow. c Western blot of SIRT2 protein in S1m tagged LncHrt vs. S1m CTRL pull down in cardiomyocytes. d Workflow of endogenous LncHrt immunoprecipitation (RIP) by SIRT2 antibody. e SIRT2 was immunoprecipitated from heart tissue and co-precipitated LncHrt were detected by qRT–PCR using 2 primers for LncHrt. IP enrichment is determined as the amount of RNA associated to SIRT2 IP relative to IgG control. Data are mean ± s.e.m. *P < 0.05 and **P < 0.01; by Student’s t test, n = 3 independent assays. f Western blot of LKB1-AMPK pathway in AAV-LncHrt injected heart tissues post myocardial infarction. g Quantification of protein expression of LKB1-AMPK pathway. Data are mean ± s.e.m. *P < 0.05 and **P < 0.01 (versus AAV-CTRL); by Student’s t test, n = 4 mice per group. h Western blot of LKB1-AMPK pathway in AAV-LncHrt shRNA injected heart tissues. i Quantification of protein expression of LKB1-AMPK pathway in AAV-LncHrt shRNA injected heart tissues. Data are mean ± s.e.m. *P < 0.05 and **P < 0.01 (versus AAV-CTRL); by Student’s t test, n = 4 mice per group. j The enrichment plot of energy dependent regulation of mTOR by LKB1-AMPK pathway by GSEA analysis
We then investigated the functional relevance of this interaction. A previous study showed that SIRT2 deacetylates downstream kinase LKB1 promoting the phosphorylation of LKB1 and subsequently activating LKB1-AMPK signaling. This pathway is cardioprotective by preserving the activity of AMPK in hypertrophic hearts [58]. As a result, we assessed whether LncHrt regulated LKB1-AMPK activation, and we found overexpression of LncHrt promoted LKB1 phosphorylation, which results downstream kinase AMPK phosphorylation at Thr172. The activation of the SIRT2 LKB1-AMPK kinase cascade by LncHrt (Fig. 6f, g) contrasts with LncHrt knock-down decreasing the phosphorylation of LKB1 and AMPK, which inhibits LKB1-AMPK signaling (Fig. 6h, i). These data are supported by our Gene Set Enrichment Analysis from the RNA-seq data of LncHrt overexpressing hearts, where energy-dependent regulation of mTOR by the LKB1-AMPK pathway is enriched (Fig. 6j). Together, these studies show that LncHrt interacts with SIRT2 to regulate its function by modulating LKB1-AMPK signaling in the infarct heart.
LncHrt preserves SIRT2 activity by blocking CDK5 inhibition
As LncHrt preserved SIRT2 activity and promoted downstream kinase activity, we asked how LncHrt impacts the SIRT2 upstream signals to positively regulate its activity. We first asked if LncHrt influences Sirt2 mRNA or SIRT2 protein levels. We established that LncHrt did not affect Sirt2 transcript levels or protein expression since Sirt2 was not changed in the LncHrt gain-of-function (Fig. 7a–c) or loss-of-function models (Fig. 7d, e).
LncHrt preserved SIRT2 activity by blocking CDK5 inhibition on SIRT2. a qRT-PCR of Sirt2 gene expression in AAV-LncHrt injected heart tissues post-MI. Rn18S as a reference gene. Data are mean ± s.e.m. *P < 0.05 and **P < 0.01 (versus AAV-CTRL); by Student’s t test, n = 4 mice per group. b Western blot of SIRT2 protein in AAV-LncHrt injected heart tissues (versus AAV-CTRL) post-MI. c Quantification of protein expression of SIRT2. Data are mean ± s.e.m. *P < 0.05 and **P < 0.01 (versus AAV-CTRL); by Student’s t test, n = 4 mice per group. d Western blot of SIRT2 protein in AAV-LncHrt KD (versus AAV-Scramble) heart tissues. e Quantification of protein expression of SIRT2. Data are mean ± s.e.m. *P < 0.05 and **P < 0.01 (versus AAV-Scramble); by Student’s t test, n = 4 mice per group. f Western blot of co-immunoprecipitated CDK5 and SIRT2 using anti-SIRT2 antibodies with negative control IgG antibody in scramble and LncHrt knock-down heart tissue lysates. g Quantification of protein enrichment of co-immunoprecipitated CDK5 in scramble and LncHrt knock-down heart tissue lysates. Data are mean ± s.e.m. *P < 0.05 and **P < 0.01; by Student’s t test, n = 3 independent assays. h Western blot of acetylated lysine and immunoprecipitated LKB1 using anti-LKB1 antibodies with negative control IgG antibody in scramble and LncHrt knock-down heart tissue lysates. i Quantification of acetylated LKB1 enrichment in scramble and LncHrt knock-down heart tissue lysates. Data are mean ± s.e.m. *P < 0.05 and **P < 0.01; by Student’s t test, n = 3 independent assays. j Western blot of LKB1-AMPK pathway in hypoxic neonatal cardiomyocytes (1% O2 treatment for 24 h) treated with CDK5 siRNA and SirReal2. k Quantification of protein expression of LKB1-AMPK pathway in hypoxic cardiomyocytes with various treatments. Data are mean ± s.e.m. *P < 0.05 and **P < 0.01; by One-way ANOVA analysis, n = 3
We then investigated the possibility that LncHrt promotes SIRT2 activity through post-translational regulation. A previous study revealed a post-translational mechanism by which p35-Cdk5 represses the catalytic activity of SIRT2 by S331 phosphorylation in post-mitotic cells [48]. Given AAV9-delivered LncHrt specifically overexpressed in cardiomyocytes, we hypothesized that CDK5 post-translationally modifies SIRT2, which suppresses LKB1-AMPK activity. We first confirmed the interaction of CDK5 with SIRT2 in the heart by co-IP (Fig. S8a). Intriguingly, we found that knock-down of LncHrt promoted the interaction between CDK5 and SIRT2 (Fig. 7f, g), and, subsequently, elevated the lysine acetylation of LKB1, resulting in abrogation of LKB1 activity and its downstream signaling cascades (Fig. 7h, i). However, LncHrt appeared to have no effect on the protein level of CDK5 in either an overexpression or depletion setting (Fig. S8, b–e). This observation ruled out the idea that LncHrt directly modulated the expression of CDK5.
Finally, we asked whether depletion of CDK5 modulates SIRT2 downstream of LKB1-AMPK signaling in cardiomyocytes. We found that knock-down of CDK5 boosted LKB1-AMPK signaling, while SIRT2 inhibition with SirReal2 effectively blocked the LKB1-AMPK activation (Fig. 7j, k). Consequently, we confirmed CDK5-mediated LKB1-AMPK signaling by modulating SIRT2 activity. LncHrt exhibited cardiac metabolic protection, at least, in part, by interfering with the catalytic activity of SIRT2, leading to LKB1-AMPK signal activation.
Therapeutic potential of LncHrt in infarcted hearts
Having demonstrated LncHrt protects the heart from MI injury, we next asked whether LncHrt could serve as potential therapeutic target to treat infarct hearts. We performed intra-myocardial injection of AAV9-LncHrt or AAV9-CTRL (2 ~ 5 × 1011 viral genome particles per mouse heart), respectively, post-MI. We carefully examined the LncHrt gene expression at 1 day, 3 days, 9 days, and 6 weeks post-MI. Similar to our previous study of using AAV-delivery [12], the expression level of LncHrt was not altered until 3 days after MI with a mild increase and appeared full expressed by 9 days post-MI and maintained for 6 weeks post-MI (i.e., when all cardiac parameters were collected) (Fig. S9a).
We observed that LncHrt improved cardiac morphology and significantly reduced the HW/BW ratio in response to MI compared to the control group (Fig. 8b). Similar to the expression pattern, we noticed no difference in cardiac function between AAV-LncHrt and AAV-CTRL injected hearts at 24 h post-MI; however, cardiac function was improved 2 weeks after LncHrt injection, as determined by echocardiography with increased fractional shortening (FS%) from MI-2 W till to MI-6 W (Fig. 8c, d and Supplementary Table 7, 8), reduced cardiac chamber internal diameter (LVID,s (mm), Fig. 8e and Supplementary Table 7, 8).
Therapeutic potential of LncHrt in infarcted hearts. a Schematic diagram showing the experimental design with AAV9-delivered LncHrt therapeutic potential in infarcted hearts. LncHrt gene expression was examined 1 day, 3 day, 9 day and 6 weeks after MI. Echocardiography was performed at post-MI 1 day, 1 week, 2 weeks, 4 weeks and 6 weeks. b Ratio of heart weight (HW) to body weight (BW) in AAV-CTRL and AAV-LncHrt heart post-MI (2 months). Data are mean ± s.e.m. *P < 0.05 and **P < 0.01 (versus AAV-CTRL); by two-way ANOVA analysis, n = 6 mice in sham group and n = 14 mice in MI group. c M-mode echocardiography of mice heart 6 weeks post-MI. d Analyses of cardiac function of FS% after AAV-LncHrt intra-myocardial injection at 1 day, 1 week, 2 weeks, 4 weeks and 6 weeks post-MI compared to their CTRL group. FS, left ventricular fractional shortening. Data are mean ± s.e.m. *P < 0.05 and **P < 0.01 (versus AAV-CTRL); by two-way ANOVA analysis, n = 6 mice in sham group and n = 17 mice in MI group. e Analyses of cardiac function of LVID after AAV-LncHrt intra-myocardial injection at 1 day, 1 week, 2 weeks, 4 weeks and 6 weeks post-MI compared to their CTRL group. LVIDs, LV internal dimension at end-systole. Data are mean ± s.e.m. *P < 0.05 and **P < 0.01 (versus AAV-CTRL); by two-way ANOVA analysis, n = 6 mice in sham group and n = 17 mice in MI group. f Representative of images of Sirius red/fast green collagen staining of series of transverse sections after injection of AAV-LncHrt compared to AAV-CTRL group after MI injury. Sirius red/fast green collagen staining marks myocardium (green) and scar (red). Scale bars = 2 mm. g Quantification of scar size of AAV-LncHrt compared to AAV-CTRL group after MI injury. Data are mean ± s.e.m. *P < 0.05 and **P < 0.01 (versus AAV-CTRL); by Student’s t test, n = 5 mice per group. h Representative images of cardiomyocytes stained with WGA. Scale bars = 100 μm. i Quantification of cardiomyocytes area of AAV-LncHrt compared to AAV-CTRL group after MI injury. Data are mean ± s.e.m. *P < 0.05, **P < 0.01 and ***P < 0.001 (versus AAV-CTRL); by two-way ANOVA analysis, n = 3 or 5 mouse hearts per group, measured ~ 200 cardiomyocytes per heart and calculated median value for each heart. j Hypertrophic marker gene expression of heart of AAV-LncHrt versus AAV-CTRL group post sham and MI. Rn18S as a reference gene. Data are mean ± s.e.m. *P < 0.05 and **P < 0.01 (versus AAV-CTRL); by two-way ANOVA analysis, n = 6 mice in sham group and n = 7 mice in MI group. k Representative respiratory experiment of mitochondrial oxidative phosphorylation (OXPHOS) capacity of complex with fatty acid oxidation pathway(F-pathway) in OXPHOS state by using application of substrate uncoupler inhibitor titration (SUIT) protocols to interrogate sequentially different substrate and coupling states using saponin permeabilized myofibers (AAV-LncHrt versus AAV- CTRL post sham and MI). l Fatty acid oxidation (FAO) after MI injury, as measured by oxygen consumption with octanoylcarnitine corrected to corresponding ROX. Data are mean ± s.e.m. *P < 0.05 and **P < 0.01 (versus AAV-CTRL post-MI); by Student’s t test, n = 9 mice per group. m Mitochondrial respiration of complex I (CI) and complex II (CII) after MI injury. The oxygen consumption rate (OCR) of complex I showed oxygen consumption with Pyruvate (P), Malate (M) and Glutamate (G) (PMG) titrations minus octanoylcarnitine titrations, while complex II respiration rate corrected to corresponding ROX. Data are mean ± s.e.m. *P < 0.05 and **P < 0.01 (versus AAV-CTRL post-MI); by Student’s t test, n = 9 mice per group. n Mitochondrial OXPHOS capacity of complex IV (CIV) after MI injury. As illustrated by OCR corrected to corresponding ROX. Data are mean ± s.e.m. *P < 0.05 and **P < 0.01 (versus AAV-CTRL post-MI); by Student’s t test, n = 9 mice per group. o Purine content in infarct heart tissues normalize to protein levels. Data are mean ± s.e.m. *P < 0.05 and **P < 0.01 (versus AAV-CTRL); by Student’s t test, n = 8 mice per group
Histological analyses revealed that LncHrt substantially reduced infarct fibrosis following MI when compared to controls (Fig. 8f, g) without induce cardiac fibrosis in sham hearts (Fig. S9. b, c). In addition, cardiomyocyte size was markedly decreased in AAV-LncHrt treated hearts post-MI, while no difference was observed in sham hearts (Fig. 8h, i). In addition, the expression of cardiac hypertrophic marker genes, Nppa, Nppb, and Myh7/Myh6 was significantly reduced in AAV-LncHrt treated hearts post-MI, while no difference was observed in sham hearts (Fig. 8j). These results indicate that intra-myocardial injection of AAV9-LncHrt prevented cardiac maladaptive remodeling post-MI.
We further assessed the therapeutic effect of LncHrt on cardiac metabolic homeostasis by measuring the oxidative phosphorylation (OXPHOS) capacity of permeabilized mouse myofibers (Fig. 8k). Respiratory stimulation of the fatty acid oxidation (FAO) pathway in the presence of malate (M) and octanoylcarnitine (Oct) was significantly elevated by LncHrt treatment post-MI (Fig. 8l), while no alteration was observed in the sham group (Fig. S9d). AAV-LncHrt also significantly promoted complex I OXPHOS capacity in the presence of pyruvate (P), glutamate (G) and malate (M), and the complex II substrate succinate oxygen consumption rate (OCR) (Fig. 8m), and tended to promote the respiration of complex IV (Fig. 8n) post-MI, while no difference was observed in sham groups (Fig. S9e and S9f). In addition, the results of purine content showed LncHrt significantly increased ATP content with decreased ADP content, indicating AAV-LncHrt treatment promotes ATP synthesis (Fig. 8o and Supplementary Table 9). However, no difference was found in the hearts of sham groups (Fig. S9g). Together, these data demonstrate that LncHrt is a potential therapeutic target for treatment of myocardial infarction.
Discussion
Here, we identified a novel cardiomyocyte-enriched lncRNA called LncHrt, which functions as an important regulator of cardiac metabolism. We found that LncHrt is the most down-regulated lncRNA in response to myocardial infarction (MI) and is essential for cardiac homeostasis, as depletion of LncHrt impairs cardiac morphology, metabolism, and function. Importantly, cardiac-specific overexpression of LncHrt protected the heart after MI through expression of genes involved in metabolic signaling and muscle contraction. These transcriptome alterations in LncHrt-overexpressing hearts compensated for MI-induced deregulation of metabolic signaling. Therefore, our study identified LncHrt as a novel cardiac lncRNA and has uncovered an unrecognized role for lncRNAs in maintaining cardiac metabolic homeostasis following pathological stress. In addition, we found the mechanism that couples lncRNAs with metabolism are mediated through the SIRT2-LKB1-AMPK axis, which provides evidence that lncRNAs can influence metabolic signaling cascades (Fig. 9).
A working model summarizing the mechanism of LncHrt function in myocardial infarction. In the infarcted heart, LncHrt is repressed through an increased interaction between CDK5 and SIRT2 accompanied by inhibition of LKB1-AMPK signaling. Under compensated LncHrt gene expression, SIRT2 activity is preserved by interfering with CDK5 inhibition of catalytic activity, then SIRT2 deacetylates LKB1 and enhances phosphorylation of LKB1, which activates LKB1-AMPK signaling. This, in turn, preserves heart function and prevents adverse cardiac remodeling by modulating metabolic homeostasis
Therapeutic strategies to attenuate adverse cardiac remodeling include increasing the supply of energy to the heart and switching the energy substrate preference to increase cardiac efficiency [14, 19]. In the infarcted heart, decreased mitochondrial respiration leads to decreased concentrations of ATP, resulting in compromised mitochondrial oxidation of fatty acids and carbohydrates, which impairs contractile function [11, 40, 44]. Promoting glucose utilization has been proposed as a means to increase ATP levels under hypoxic conditions [27]; however, decreased fatty acid oxidation is considered maladaptive [28, 54]. On the other hand, high fat diet-fed rats subjected to MI showed improved cardiac function through enhance fatty acid oxidation [2]. So, this metabolic phenotype affects recovery of the injured heart [2, 9]. With that in mind, we found that LncHrt not only accelerates glucose utilization in the infarcted myocardium, but also enhances mitochondrial respiration in intact cardiomyocytes from the non-infarcted area, which indicates LncHrt improves cardiac metabolism and protects the heart from ischemia.
During MI, AMPK is activated to maintain energy by increasing glycolysis and accelerating fatty acid oxidation [17]. Phosphorylation at Thr172 activates AMPK activity [69], which is mediated through LKB1 [67]. Deacetylation of LKB1 influences its intracellular localization, association with its partner protein STRAD, kinase activity, and its ability to activate AMPK [33]. Our study shows MI-induced downregulation of LncHrt. By overexpressing this lncRNA post-MI, AMPK activity can be increased to protect the ischemic heart. Therefore, LncHrt may function as an energy sensor.
The cyclin-dependent kinase CDK5 is important for neuronal development, survival, migration, differentiation, and metabolism [71]. CDK5 phosphorylates AMPKa2 at Thr485, which repressed its activity in the brain [39]. While we did not observe an interaction between CDK5 and AMPKa2 in the heart, CDK5 inhibition may be cardioprotective [24]. Suppressing CDK5 activity also alleviates the cardiac phenotype associated with Timothy Syndrome [55]. Although, we have shown that inhibition of CDK5 positively regulates LKB1 and AMPK activity, the pathophysiological role of this kinase has yet to be fully determined.
SIRT2 is the only sirtuin that is preferentially localized to the cytoplasm. SIRT2 plays an essential role in many aspects of metabolism [36, 58, 66, 72]. In particular, SIRT2 restrains cardiac hypertrophy by deacetylating LKB1 and activating LKB1-AMPK signaling [58]. CDK5 phosphorylates Ser331 in the C-terminal portion of the catalytic domain of SIRT2 to inhibit deacetylase activity of this protein, which affects neuronal adhesion, migration, and outgrowth [48]. In our study, we established that LncHrt regulated LKB1-AMPK activity by interfering with the CDK5/SIRT2 interaction and modulating cardiac metabolism. We speculate that manipulation of CDK5 activity has therapeutic potential for heart disease.
In this study, we demonstrated that LncHrt plays an important role in cardiac metabolic homeostasis post-MI through regulation of (1) metabolic gene expression; (2) mitochondrial respiration and (3) SIRT2-LKB1-AMPK signaling. However, we have not revealed these 3 LncHrt-modulated activities are connected and to what extent they contribute to cardioprotection after MI. Given that many studies have shown direct or indirect transcriptional regulation by AMPK, we speculate that upon LncHrt-stimulation of the SIRT2-LKB1-AMPK axis post-MI, activated AMPK controls metabolic gene expression through metabolites of intermediary metabolism or by modulating transcription factors and cofactors, such as CREB1 [6], PGC-1α, FOXO1 [4], and others [56]. Presumably, LncHrt-activated AMPK could transcriptionally modulate gene expression of metabolism and control energy homeostasis in cardiac disease. In addition, it is unclear whether LncHrt-mediated activation of the SIRT2-LKB1-AMPK axis is required to preserve mitochondrial respiration and/or prevent maladaptive remodeling after MI. As a result, a more complete view of LncHrt-regulated metabolic homeostasis (including signaling, mitochondrial respiration and gene expression) needs to be elucidated.
Most LncRNAs lack sequence conservation across species and the secondary structure of lncRNAs is more highly conserved than the RNA sequence [49, 59, 65]. However, we identified the gene locus of LncHrt in the human genome that is positionally conserved (i.e., both reside on chromosome 14) and conserved with respect to its sequence (i.e., in exon 1 and portions of exon 2 across multiple species). Combined analysis with MFE predicted the secondary structure of LncHrt with potential base-paring sites are also conserved. Furthermore, the expression of the human homolog of LncHrt is downregulated in patients with dilated cardiomyopathy. In view of that, we evaluated the therapeutic efficacy of AAV9-delivered LncHrt in mice as the effectiveness and safety of AAV-directed gene therapy has been proven in patients [42]. From this perspective, AAV9-delivery of LncHrt may represent a novel RNA-based therapy for ischemic heart disease.
References
Bak MI, Ingwall JS (1994) Acidosis during ischemia promotes adenosine triphosphate resynthesis in postischemic rat heart. In vivo regulation of 5’-nucleotidase. J Clin Invest 93:40–49. https://doi.org/10.1172/JCI116974
Berthiaume JM, Young ME, Chen X, McElfresh TA, Yu X, Chandler MP (2012) Normalizing the metabolic phenotype after myocardial infarction: impact of subchronic high fat feeding. J Mol Cell Cardiol 53:125–133. https://doi.org/10.1016/j.yjmcc.2012.04.005
Cai B, Ma W, Ding F, Zhang L, Huang Q, Wang X, Hua B, Xu J, Li J, Bi C, Guo S, Yang F, Han Z, Li Y, Yan G, Yu Y, Bao Z, Yu M, Li F, Tian Y, Pan Z, Yang B (2018) The long noncoding RNA CAREL controls cardiac regeneration. J Am Coll Cardiol 72:534–550. https://doi.org/10.1016/j.jacc.2018.04.085
Canto C, Jiang LQ, Deshmukh AS, Mataki C, Coste A, Lagouge M, Zierath JR, Auwerx J (2010) Interdependence of AMPK and SIRT1 for metabolic adaptation to fasting and exercise in skeletal muscle. Cell Metab 11:213–219. https://doi.org/10.1016/j.cmet.2010.02.006
Chen J, Shearer GC, Chen Q, Healy CL, Beyer AJ, Nareddy VB, Gerdes AM, Harris WS, O’Connell TD, Wang D (2011) Omega-3 fatty acids prevent pressure overload-induced cardiac fibrosis through activation of cyclic GMP/protein kinase G signaling in cardiac fibroblasts. Circulation 123:584–593. https://doi.org/10.1161/CIRCULATIONAHA.110.971853
Chhipa RR, Fan Q, Anderson J, Muraleedharan R, Huang Y, Ciraolo G, Chen X, Waclaw R, Chow LM, Khuchua Z, Kofron M, Weirauch MT, Kendler A, McPherson C, Ratner N, Nakano I, Dasgupta N, Komurov K, Dasgupta B (2018) AMP kinase promotes glioblastoma bioenergetics and tumour growth. Nat Cell Biol 20:823–835. https://doi.org/10.1038/s41556-018-0126-z
Cohn JN, Ferrari R, Sharpe N (2000) Cardiac remodeling–concepts and clinical implications: a consensus paper from an international forum on cardiac remodeling. Behalf of an international forum on cardiac remodeling. J Am Coll Cardiol 35:569–582. https://doi.org/10.1016/s0735-1097(99)00630-0
de Castro Bras LE, Cates CA, DeLeon-Pennell KY, Ma Y, Iyer RP, Halade GV, Yabluchanskiy A, Fields GB, Weintraub ST, Lindsey ML (2014) Citrate synthase is a novel in vivo matrix metalloproteinase-9 substrate that regulates mitochondrial function in the postmyocardial infarction left ventricle. Antioxid Redox Signal 21:1974–1985. https://doi.org/10.1089/ars.2013.5411
Diebold S, Moellmann J, Kahles F, Haj-Yehia E, Liehn EA, Nickel A, Lebherz C, Maack C, Marx N, Lehrke M (2018) Myocardial infarction is sufficient to increase GLP-1 secretion, leading to improved left ventricular contractility and mitochondrial respiratory capacity. Diabetes Obes Metab 20:2911–2918. https://doi.org/10.1111/dom.13472
Ding J, Chen J, Wang Y, Kataoka M, Ma L, Zhou P, Hu X, Lin Z, Nie M, Deng ZL, Pu WT, Wang DZ (2015) Trbp regulates heart function through microRNA-mediated Sox6 repression. Nat Genet 47:776–783. https://doi.org/10.1038/ng.3324
Galan DT, Bito V, Claus P, Holemans P, Abi-Char J, Nagaraju CK, Dries E, Vermeulen K, Ventura-Clapier R, Sipido KR, Driesen RB (2016) Reduced mitochondrial respiration in the ischemic as well as in the remote nonischemic region in postmyocardial infarction remodeling. Am J Physiol Heart Circ Physiol 311:H1075–H1090. https://doi.org/10.1152/ajpheart.00945.2015
Gao F, Kataoka M, Liu N, Liang T, Huang ZP, Gu F, Ding J, Liu J, Zhang F, Ma Q, Wang Y, Zhang M, Hu X, Kyselovic J, Hu X, Pu WT, Wang J, Chen J, Wang DZ (2019) Therapeutic role of miR-19a/19b in cardiac regeneration and protection from myocardial infarction. Nat Commun 10:1802. https://doi.org/10.1038/s41467-019-09530-1
Gao L, Wang X, Guo S, Xiao L, Liang C, Wang Z, Li Y, Liu Y, Yao R, Liu Y, Zhang Y (2019) LncRNA HOTAIR functions as a competing endogenous RNA to upregulate SIRT1 by sponging miR-34a in diabetic cardiomyopathy. J Cell Physiol 234:4944–4958. https://doi.org/10.1002/jcp.27296
Gibb AA, Hill BG (2018) Metabolic coordination of physiological and pathological cardiac remodeling. Circ Res 123:107–128. https://doi.org/10.1161/CIRCRESAHA.118.312017
Grieger JC, Choi VW, Samulski RJ (2006) Production and characterization of adeno-associated viral vectors. Nat Protoc 1:1412–1428. https://doi.org/10.1038/nprot.2006.207
Griffin LD, Gelb BD, Adams V, McCabe ER (1992) Developmental expression of hexokinase 1 in the rat. Biochim Biophys Acta 1129:309–317. https://doi.org/10.1016/0167-4781(92)90508-w
Gundewar S, Calvert JW, Jha S, Toedt-Pingel I, Ji SY, Nunez D, Ramachandran A, Anaya-Cisneros M, Tian R, Lefer DJ (2009) Activation of AMP-activated protein kinase by metformin improves left ventricular function and survival in heart failure. Circ Res 104:403–411. https://doi.org/10.1161/CIRCRESAHA.108.190918
Liu H, J Y, Luo Y, Jiang W (2006) A simple and rapid determination of ATP, ADP and AMP concentrations in pericarp tissue of litchi fruit by high performance liquid chromatography. Food Technol Biotechnol 44:531–534
Heusch G, Libby P, Gersh B, Yellon D, Bohm M, Lopaschuk G, Opie L (2014) Cardiovascular remodelling in coronary artery disease and heart failure. Lancet 383:1933–1943. https://doi.org/10.1016/S0140-6736(14)60107-0
Huang ZP, Ding Y, Chen J, Wu G, Kataoka M, Hu Y, Yang JH, Liu J, Drakos SG, Selzman CH, Kyselovic J, Qu LH, Dos Remedios CG, Pu WT, Wang DZ (2016) Long non-coding RNAs link extracellular matrix gene expression to ischemic cardiomyopathy. Cardiovasc Res. https://doi.org/10.1093/cvr/cvw201
Huang ZP, Kataoka M, Chen J, Wu G, Ding J, Nie M, Lin Z, Liu J, Hu X, Ma L, Zhou B, Wakimoto H, Zeng C, Kyselovic J, Deng ZL, Seidman CE, Seidman JG, Pu WT, Wang DZ (2015) Cardiomyocyte-enriched protein CIP protects against pathophysiological stresses and regulates cardiac homeostasis. J Clin Invest 125:4122–4134. https://doi.org/10.1172/JCI82423
Ingwall JS (2009) Energy metabolism in heart failure and remodelling. Cardiovasc Res 81:412–419. https://doi.org/10.1093/cvr/cvn301
Jaenisch RB, Bertagnolli M, Borghi-Silva A, Arena R, Lago PD (2017) Respiratory muscle training improves diaphragm citrate synthase activity and hemodynamic function in rats with heart failure. Braz J Cardiovasc Surg 32:104–110. https://doi.org/10.21470/1678-9741-2017-0002
Jungk L, Franke H, Salameh A, Dhein S (2019) Golgi fragmentation in human patients with chronic atrial fibrillation: a new aspect of remodeling. Thorac Cardiovasc Surg 67:98–106. https://doi.org/10.1055/s-0038-1635079
Karwi QG, Uddin GM, Ho KL, Lopaschuk GD (2018) Loss of metabolic flexibility in the failing heart. Front Cardiovasc Med 5:68. https://doi.org/10.3389/fcvm.2018.00068
Kim AS, Miller EJ, Wright TM, Li J, Qi D, Atsina K, Zaha V, Sakamoto K, Young LH (2011) A small molecule AMPK activator protects the heart against ischemia-reperfusion injury. J Mol Cell Cardiol 51:24–32. https://doi.org/10.1016/j.yjmcc.2011.03.003
King LM, Opie LH (1998) Glucose and glycogen utilisation in myocardial ischemia—changes in metabolism and consequences for the myocyte. Mol Cell Biochem 180:3–26
Kolwicz SC Jr, Purohit S, Tian R (2013) Cardiac metabolism and its interactions with contraction, growth, and survival of cardiomyocytes. Circ Res 113:603–616. https://doi.org/10.1161/CIRCRESAHA.113.302095
Kolwicz SC Jr, Tian R (2011) Glucose metabolism and cardiac hypertrophy. Cardiovasc Res 90:194–201. https://doi.org/10.1093/cvr/cvr071
Kong L, Zhang Y, Ye ZQ, Liu XQ, Zhao SQ, Wei L, Gao G (2007) CPC: assess the protein-coding potential of transcripts using sequence features and support vector machine. Nucleic Acids Res 35:W345-349. https://doi.org/10.1093/nar/gkm391
Ku HJ, Ahn Y, Lee JH, Park KM, Park JW (2015) IDH2 deficiency promotes mitochondrial dysfunction and cardiac hypertrophy in mice. Free Radic Biol Med 80:84–92. https://doi.org/10.1016/j.freeradbiomed.2014.12.018
Kurian L, Aguirre A, Sancho-Martinez I, Benner C, Hishida T, Nguyen TB, Reddy P, Nivet E, Krause MN, Nelles DA, Esteban CR, Campistol JM, Yeo GW, Belmonte JCI (2015) Identification of novel long noncoding RNAs underlying vertebrate cardiovascular development. Circulation 131:1278–1290. https://doi.org/10.1161/CIRCULATIONAHA.114.013303
Lan F, Cacicedo JM, Ruderman N, Ido Y (2008) SIRT1 modulation of the acetylation status, cytosolic localization, and activity of LKB1. Possible role in AMP-activated protein kinase activation. J Biol Chem 283:27628–27635. https://doi.org/10.1074/jbc.M805711200
Lemieux H, Blier PU, Gnaiger E (2017) Remodeling pathway control of mitochondrial respiratory capacity by temperature in mouse heart: electron flow through the Q-junction in permeabilized fibers. Sci Rep 7:2840. https://doi.org/10.1038/s41598-017-02789-8
Lemieux H, Semsroth S, Antretter H, Hofer D, Gnaiger E (2011) Mitochondrial respiratory control and early defects of oxidative phosphorylation in the failing human heart. Int J Biochem Cell Biol 43:1729–1738. https://doi.org/10.1016/j.biocel.2011.08.008
Lemos V, de Oliveira RM, Naia L, Szego E, Ramos E, Pinho S, Magro F, Cavadas C, Rego AC, Costa V, Outeiro TF, Gomes P (2017) The NAD+-dependent deacetylase SIRT2 attenuates oxidative stress and mitochondrial dysfunction and improves insulin sensitivity in hepatocytes. Hum Mol Genet 26:4105–4117. https://doi.org/10.1093/hmg/ddx298
Leppek K, Stoecklin G (2014) An optimized streptavidin-binding RNA aptamer for purification of ribonucleoprotein complexes identifies novel ARE-binding proteins. Nucleic Acids Res 42:e13. https://doi.org/10.1093/nar/gkt956
Li X, Jiang Y, Meisenhelder J, Yang W, Hawke DH, Zheng Y, Xia Y, Aldape K, He J, Hunter T, Wang L, Lu Z (2016) Mitochondria-translocated PGK1 functions as a protein kinase to coordinate glycolysis and the TCA Cycle in tumorigenesis. Mol Cell 61:705–719. https://doi.org/10.1016/j.molcel.2016.02.009
Li Y, Xiang Q, Yao YH, Li JJ, Wang Y, Li XH (2020) Inactivated AMPK-alpha2 promotes the progression of diabetic brain damage by Cdk5 phosphorylation at Thr485 site. Biochimie 168:277–284. https://doi.org/10.1016/j.biochi.2019.11.010
Lopaschuk GD, Ussher JR, Folmes CD, Jaswal JS, Stanley WC (2010) Myocardial fatty acid metabolism in health and disease. Physiol Rev 90:207–258. https://doi.org/10.1152/physrev.00015.2009
Makrecka-Kuka M, Krumschnabel G, Gnaiger E (2015) High-resolution respirometry for simultaneous measurement of oxygen and hydrogen peroxide fluxes in permeabilized cells, tissue homogenate and isolated mitochondria. Biomolecules 5:1319–1338. https://doi.org/10.3390/biom5031319
Mendell JR, Al-Zaidy S, Shell R, Arnold WD, Rodino-Klapac LR, Prior TW, Lowes L, Alfano L, Berry K, Church K, Kissel JT, Nagendran S, L’Italien J, Sproule DM, Wells C, Cardenas JA, Heitzer MD, Kaspar A, Corcoran S, Braun L, Likhite S, Miranda C, Meyer K, Foust KD, Burghes AHM, Kaspar BK (2017) Single-dose gene-replacement therapy for spinal muscular atrophy. N Engl J Med 377:1713–1722. https://doi.org/10.1056/NEJMoa1706198
Micheletti R, Plaisance I, Abraham BJ, Sarre A, Ting CC, Alexanian M, Maric D, Maison D, Nemir M, Young RA, Schroen B, Gonzalez A, Ounzain S, Pedrazzini T (2017) The long noncoding RNA Wisper controls cardiac fibrosis and remodeling. Sci Transl Med. https://doi.org/10.1126/scitranslmed.aai9118
Nascimben L, Friedrich J, Liao R, Pauletto P, Pessina AC, Ingwall JS (1995) Enalapril treatment increases cardiac performance and energy reserve via the creatine kinase reaction in myocardium of Syrian myopathic hamsters with advanced heart failure. Circulation 91:1824–1833. https://doi.org/10.1161/01.cir.91.6.1824
Neubauer S, Horn M, Naumann A, Tian R, Hu K, Laser M, Friedrich J, Gaudron P, Schnackerz K, Ingwall JS et al (1995) Impairment of energy metabolism in intact residual myocardium of rat hearts with chronic myocardial infarction. J Clin Invest 95:1092–1100. https://doi.org/10.1172/JCI117756
Oliver MF (1972) Metabolic response during impending myocardial infarction. II. Clinical implications. Circulation 45:491–500. https://doi.org/10.1161/01.cir.45.2.491
Ounzain S, Micheletti R, Beckmann T, Schroen B, Alexanian M, Pezzuto I, Crippa S, Nemir M, Sarre A, Johnson R, Dauvillier J, Burdet F, Ibberson M, Guigo R, Xenarios I, Heymans S, Pedrazzini T (2015) Genome-wide profiling of the cardiac transcriptome after myocardial infarction identifies novel heart-specific long non-coding RNAs. Eur Heart J 36:353–368a. https://doi.org/10.1093/eurheartj/ehu180
Pandithage R, Lilischkis R, Harting K, Wolf A, Jedamzik B, Luscher-Firzlaff J, Vervoorts J, Lasonder E, Kremmer E, Knoll B, Luscher B (2008) The regulation of SIRT2 function by cyclin-dependent kinases affects cell motility. J Cell Biol 180:915–929. https://doi.org/10.1083/jcb.200707126
Pang KC, Frith MC, Mattick JS (2006) Rapid evolution of noncoding RNAs: lack of conservation does not mean lack of function. Trends Genet 22:1–5. https://doi.org/10.1016/j.tig.2005.10.003
Pfeffer JM, Pfeffer MA, Fletcher PJ, Braunwald E (1991) Progressive ventricular remodeling in rat with myocardial infarction. Am J Physiol 260:H1406-1414. https://doi.org/10.1152/ajpheart.1991.260.5.H1406
Roth GA, Johnson C, Abajobir A, Abd-Allah F, Abera SF, Abyu G, Ahmed M, Aksut B, Alam T, Alam K, Alla F, Alvis-Guzman N, Amrock S, Ansari H, Arnlov J, Asayesh H, Atey TM, Avila-Burgos L, Awasthi A, Banerjee A, Barac A, Barnighausen T, Barregard L, Bedi N, Belay Ketema E, Bennett D, Berhe G, Bhutta Z, Bitew S, Carapetis J, Carrero JJ, Malta DC, Castaneda-Orjuela CA, Castillo-Rivas J, Catala-Lopez F, Choi JY, Christensen H, Cirillo M, Cooper L Jr, Criqui M, Cundiff D, Damasceno A, Dandona L, Dandona R, Davletov K, Dharmaratne S, Dorairaj P, Dubey M, Ehrenkranz R, El Sayed ZM, Faraon EJA, Esteghamati A, Farid T, Farvid M, Feigin V, Ding EL, Fowkes G, Gebrehiwot T, Gillum R, Gold A, Gona P, Gupta R, Habtewold TD, Hafezi-Nejad N, Hailu T, Hailu GB, Hankey G, Hassen HY, Abate KH, Havmoeller R, Hay SI, Horino M, Hotez PJ, Jacobsen K, James S, Javanbakht M, Jeemon P, John D, Jonas J, Kalkonde Y, Karimkhani C, Kasaeian A, Khader Y, Khan A, Khang YH, Khera S, Khoja AT, Khubchandani J, Kim D, Kolte D, Kosen S, Krohn KJ, Kumar GA, Kwan GF, Lal DK, Larsson A, Linn S, Lopez A, Lotufo PA, El Razek HMA et al (2017) Global, regional, and national burden of cardiovascular diseases for 10 causes, 1990 to 2015. J Am Coll Cardiol 70:1–25. https://doi.org/10.1016/j.jacc.2017.04.052
Russell RR 3rd, Li J, Coven DL, Pypaert M, Zechner C, Palmeri M, Giordano FJ, Mu J, Birnbaum MJ, Young LH (2004) AMP-activated protein kinase mediates ischemic glucose uptake and prevents postischemic cardiac dysfunction, apoptosis, and injury. J Clin Invest 114:495–503. https://doi.org/10.1172/JCI19297
Sarikhani M, Maity S, Mishra S, Jain A, Tamta AK, Ravi V, Kondapalli MS, Desingu PA, Khan D, Kumar S, Rao S, Inbaraj M, Pandit AS, Sundaresan NR (2018) SIRT2 deacetylase represses NFAT transcription factor to maintain cardiac homeostasis. J Biol Chem 293:5281–5294. https://doi.org/10.1074/jbc.RA117.000915
Sena S, Hu P, Zhang D, Wang X, Wayment B, Olsen C, Avelar E, Abel ED, Litwin SE (2009) Impaired insulin signaling accelerates cardiac mitochondrial dysfunction after myocardial infarction. J Mol Cell Cardiol 46:910–918. https://doi.org/10.1016/j.yjmcc.2009.02.014
Song L, Park SE, Isseroff Y, Morikawa K, Yazawa M (2017) Inhibition of CDK5 alleviates the cardiac phenotypes in timothy syndrome. Stem Cell Rep 9:50–57. https://doi.org/10.1016/j.stemcr.2017.05.028
Sukumaran A, Choi K, Dasgupta B (2020) Insight on transcriptional regulation of the energy sensing AMPK and biosynthetic mTOR pathway genes. Front Cell Dev Biol 8:671. https://doi.org/10.3389/fcell.2020.00671
Sun D, Yang F (2017) Metformin improves cardiac function in mice with heart failure after myocardial infarction by regulating mitochondrial energy metabolism. Biochem Biophys Res Commun 486:329–335. https://doi.org/10.1016/j.bbrc.2017.03.036
Tang X, Chen XF, Wang NY, Wang XM, Liang ST, Zheng W, Lu YB, Zhao X, Hao DL, Zhang ZQ, Zou MH, Liu DP, Chen HZ (2017) SIRT2 acts as a cardioprotective deacetylase in pathological cardiac hypertrophy. Circulation 136:2051–2067. https://doi.org/10.1161/CIRCULATIONAHA.117.028728
Torarinsson E, Sawera M, Havgaard JH, Fredholm M, Gorodkin J (2006) Thousands of corresponding human and mouse genomic regions unalignable in primary sequence contain common RNA structure. Genome Res 16:885–889. https://doi.org/10.1101/gr.5226606
van Rooij E (2016) Cardiac repair after myocardial infarction. N Engl J Med 374:85–87. https://doi.org/10.1056/NEJMcibr1512011
Veksler VI, Kuznetsov AV, Sharov VG, Kapelko VI, Saks VA (1987) Mitochondrial respiratory parameters in cardiac tissue: a novel method of assessment by using saponin-skinned fibers. Biochim Biophys Acta 892:191–196. https://doi.org/10.1016/0005-2728(87)90174-5
Viereck J, Kumarswamy R, Foinquinos A, Xiao K, Avramopoulos P, Kunz M, Dittrich M, Maetzig T, Zimmer K, Remke J, Just A, Fendrich J, Scherf K, Bolesani E, Schambach A, Weidemann F, Zweigerdt R, de Windt LJ, Engelhardt S, Dandekar T, Batkai S, Thum T (2016) Long noncoding RNA Chast promotes cardiac remodeling. Sci Transl Med 8:3260322. https://doi.org/10.1126/scitranslmed.aaf1475
Wang L, Park HJ, Dasari S, Wang S, Kocher JP, Li W (2013) CPAT: Coding-potential assessment tool using an alignment-free logistic regression model. Nucleic Acids Res 41:e74. https://doi.org/10.1093/nar/gkt006
Wang Z, Zhang XJ, Ji YX, Zhang P, Deng KQ, Gong J, Ren S, Wang X, Chen I, Wang H, Gao C, Yokota T, Ang YS, Li S, Cass A, Vondriska TM, Li G, Deb A, Srivastava D, Yang HT, Xiao X, Li H, Wang Y (2016) The long noncoding RNA Chaer defines an epigenetic checkpoint in cardiac hypertrophy. Nat Med 22:1131–1139. https://doi.org/10.1038/nm.4179
Washietl S, Hofacker IL, Lukasser M, Huttenhofer A, Stadler PF (2005) Mapping of conserved RNA secondary structures predicts thousands of functional noncoding RNAs in the human genome. Nat Biotechnol 23:1383–1390. https://doi.org/10.1038/nbt1144
Watanabe H, Inaba Y, Kimura K, Matsumoto M, Kaneko S, Kasuga M, Inoue H (2018) Sirt2 facilitates hepatic glucose uptake by deacetylating glucokinase regulatory protein. Nat Commun 9:30. https://doi.org/10.1038/s41467-017-02537-6
Woods A, Johnstone SR, Dickerson K, Leiper FC, Fryer LG, Neumann D, Schlattner U, Wallimann T, Carlson M, Carling D (2003) LKB1 is the upstream kinase in the AMP-activated protein kinase cascade. Curr Biol 13:2004–2008. https://doi.org/10.1016/j.cub.2003.10.031
Wu G, Cai J, Han Y, Chen J, Huang ZP, Chen C, Cai Y, Huang H, Yang Y, Liu Y, Xu Z, He D, Zhang X, Hu X, Pinello L, Zhong D, He F, Yuan GC, Wang DZ, Zeng C (2014) LincRNA-p21 regulates neointima formation, vascular smooth muscle cell proliferation, apoptosis, and atherosclerosis by enhancing p53 activity. Circulation 130:1452–1465. https://doi.org/10.1161/CIRCULATIONAHA.114.011675
Xiao B, Sanders MJ, Underwood E, Heath R, Mayer FV, Carmena D, Jing C, Walker PA, Eccleston JF, Haire LF, Saiu P, Howell SA, Aasland R, Martin SR, Carling D, Gamblin SJ (2011) Structure of mammalian AMPK and its regulation by ADP. Nature 472:230–233. https://doi.org/10.1038/nature09932
Xiao C, Wang K, Xu Y, Hu H, Zhang N, Wang Y, Zhong Z, Zhao J, Li Q, Zhu D, Ke C, Zhong S, Wu X, Yu H, Zhu W, Chen J, Zhang J, Wang J, Hu X (2018) Transplanted mesenchymal stem cells reduce autophagic flux in infarcted hearts via the exosomal transfer of miR-125b. Circ Res 123:564–578. https://doi.org/10.1161/CIRCRESAHA.118.312758
Xie Z, Sanada K, Samuels BA, Shih H, Tsai LH (2003) Serine 732 phosphorylation of FAK by Cdk5 is important for microtubule organization, nuclear movement, and neuronal migration. Cell 114:469–482. https://doi.org/10.1016/s0092-8674(03)00605-6
Yang W, Gao F, Zhang P, Pang S, Cui Y, Liu L, Wei G, Yan B (2017) Functional genetic variants within the SIRT2 gene promoter in acute myocardial infarction. PLoS One 12:e0176245. https://doi.org/10.1371/journal.pone.0176245
Zhang F, Fu X, Kataoka M, Liu N, Wang Y, Gao F, Liang T, Dong X, Pei J, Hu X, Zhu W, Yu H, Cowan DB, Hu X, Huang ZP, Wang J, Wang DZ, Chen J (2021) Long noncoding RNA Cfast regulates cardiac fibrosis. Mol Ther Nucleic Acids 23:377–392. https://doi.org/10.1016/j.omtn.2020.11.013
Zornoff LA, Paiva SA, Duarte DR, Spadaro J (2009) Ventricular remodeling after myocardial infarction: concepts and clinical implications. Arq Bras Cardiol 92:150–164. https://doi.org/10.1590/s0066-782x2009000200013
Acknowledgements
This work is supported by National Key R&D Program of China (2017YFA0103700); Zhejiang Provincial NSF project (LZ20H020001 to J. Chen.); National Natural Science Foundation of China (Nos. 81470382, 81670257, 81970227 to J. Chen, and 82000244 to F. Gao,); China Postdoctoral Science Foundation (2020M671751 and 2021T140596 to F. Gao). Work in the Wang laboratory is supported by NIH grant HL125925.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
On behalf of all authors, the corresponding author states that there is no conflict of interest.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Liu, N., Kataoka, M., Wang, Y. et al. LncRNA LncHrt preserves cardiac metabolic homeostasis and heart function by modulating the LKB1-AMPK signaling pathway. Basic Res Cardiol 116, 48 (2021). https://doi.org/10.1007/s00395-021-00887-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00395-021-00887-3