Abstract
Purpose
While conventional statistical approaches have been used to identify risk factors for cerebrospinal fluid (CSF) shunt failure, these methods may not fully capture the complex contribution of clinical, radiologic, surgical, and shunt-specific variables influencing this outcome. Using prospectively collected data from the Hydrocephalus Clinical Research Network (HCRN) patient registry, we applied machine learning (ML) approaches to create a predictive model of CSF shunt failure.
Methods
Pediatric patients (age < 19 years) undergoing first-time CSF shunt placement at six HCRN centers were included. CSF shunt failure was defined as a composite outcome including requirement for shunt revision, endoscopic third ventriculostomy, or shunt infection within 5 years of initial surgery. Performance of conventional statistical and 4 ML models were compared.
Results
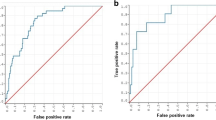
Our cohort consisted of 1036 children undergoing CSF shunt placement, of whom 344 (33.2%) experienced shunt failure. Thirty-eight clinical, radiologic, surgical, and shunt-design variables were included in the ML analyses. Of all ML algorithms tested, the artificial neural network (ANN) had the strongest performance with an area under the receiver operator curve (AUC) of 0.71. The ANN had a specificity of 90% and a sensitivity of 68%, meaning that the ANN can effectively rule-in patients most likely to experience CSF shunt failure (i.e., high specificity) and moderately effective as a tool to rule-out patients at high risk of CSF shunt failure (i.e., moderately sensitive). The ANN was independently validated in 155 patients (prospectively collected, retrospectively analyzed).
Conclusion
These data suggest that the ANN, or future iterations thereof, can provide an evidence-based tool to assist in prognostication and patient-counseling immediately after CSF shunt placement.

Similar content being viewed by others
References
Lim J, Tang AR, Liles C, Hysong AA, Hale AT, Bonfield CM, Naftel RP, Wellons JC, Shannon CN (2018) The cost of hydrocephalus: a cost-effectiveness model for evaluating surgical techniques 1
Lazareff JA, Peacock W, Holly L, Ver Halen J, Wong A, Olmstead C (1998) Multiple shunt failures: an analysis of relevant factors. Childs Nerv Syst 14:271–275
Tuli S, Drake J, Lawless J, Wigg M, Lamberti-Pasculli M (2000) Risk factors for repeated cerebrospinal shunt failures in pediatric patients with hydrocephalus. J Neurosurg 92:31–38
Riva-Cambrin J, Kestle JR, Holubkov R, Butler J, Kulkarni AV, Drake J, Whitehead WE, Wellons JC 3rd, Shannon CN, Tamber MS, Limbrick DD Jr, Rozzelle C, Browd SR, Simon TD (2016) Risk factors for shunt malfunction in pediatric hydrocephalus: a multicenter prospective cohort study. J Neurosurg Pediatr 17:382–390
Tomycz LD, Hale AT, George TM (2017) Emerging insights and new perspectives on the nature of hydrocephalus. Pediatr Neurosurg 52:361–368
Rajkomar A, Dean J, Kohane I (2019) Machine learning in medicine. N Engl J Med 380:1347–1358
Buchlak QD, Esmaili N, Leveque JC, Farrokhi F, Bennett C, Piccardi M, Sethi RK (2019) Machine learning applications to clinical decision support in neurosurgery: an artificial intelligence augmented systematic review. Neurosurg Rev
Senders JT, Staples PC, Karhade AV, Zaki MM, Gormley WB, Broekman MLD, Smith TR, Arnaout O (2018) Machine learning and neurosurgical outcome prediction: a systematic review. World Neurosurg 109:476–486.e471
Cherukuri V, Ssenyonga P, Warf BC, Kulkarni AV, Monga V, Schiff SJ (2018) Learning based segmentation of CT brain images: application to postoperative hydrocephalic scans. IEEE Trans Biomed Eng 65:1871–1884
Hale AT, Stonko DP, Brown A, Lim J, Voce DJ, Gannon SR, Le TM, Shannon CN (2018) Machine-learning analysis outperforms conventional statistical models and CT classification systems in predicting 6-month outcomes in pediatric patients sustaining traumatic brain injury. Neurosurg Focus 45:E2
Hale AT, Stonko DP, Lim J, Guillamondegui OD, Shannon CN, Patel MB (2018) Using an artificial neural network to predict traumatic brain injury. J Neurosurg Pediatr:1–8
Hale AT, Stonko DP, Wang L, Strother MK, Chambless LB (2018) Machine learning analyses can differentiate meningioma grade by features on magnetic resonance imaging. Neurosurg Focus 45:E4
Pisapia JM, Akbari H, Rozycki M, Goldstein H, Bakas S, Rathore S, Moldenhauer JS, Storm PB, Zarnow DM, Anderson RCE, Heuer GG, Davatzikos C (2018) Use of fetal magnetic resonance image analysis and machine learning to predict the need for postnatal cerebrospinal fluid diversion in fetal ventriculomegaly. JAMA Pediatr 172:128–135
Darcy AM, Louie AK, Roberts LW (2016) Machine learning and the profession of medicine. JAMA 315:551–552
Obermeyer Z, Emanuel EJ (2016) Predicting the future - big data, machine learning, and clinical medicine. N Engl J Med 375:1216–1219
Kestle JR, Riva-Cambrin J, Wellons JC 3rd, Kulkarni AV, Whitehead WE, Walker ML, Oakes WJ, Drake JM, Luerssen TG, Simon TD, Holubkov R (2011) A standardized protocol to reduce cerebrospinal fluid shunt infection: the Hydrocephalus Clinical Research Network Quality Improvement Initiative. J Neurosurg Pediatr 8:22–29
Simon TD, Butler J, Whitlock KB, Browd SR, Holubkov R, Kestle JR, Kulkarni AV, Langley M, Limbrick DD Jr, Mayer-Hamblett N, Tamber M, Wellons JC 3rd, Whitehead WE, Riva-Cambrin J (2014) Risk factors for first cerebrospinal fluid shunt infection: findings from a multi-center prospective cohort study. J Pediatr 164:1462–1468.e1462
Gestel TV, Suykens JAK, Lanckriet G, Lambrechts A, Moor BD, Vandewalle J (2002) Bayesian framework for least-squares support vector machine classifiers, Gaussian processes, and kernel Fisher discriminant analysis. Neural Comput 14:1115–1147
Møller MF (1993) A scaled conjugate gradient algorithm for fast supervised learning. Neural Netw 6:525–533
Kulkarni AV, Drake JM, Armstrong DC, Dirks PB (1999) Measurement of ventricular size: reliability of the frontal and occipital horn ratio compared to subjective assessment. Pediatr Neurosurg 31:65–70
Harbaugh RE (2018) Editorial. Artificial neural networks for neurosurgical diagnosis, prognosis, and management. Neurosurg Focus 45:E3
Rossi NB, Khan NR, Jones TL, Lepard J, McAbee JH, Klimo P Jr (2016) Predicting shunt failure in children: should the global shunt revision rate be a quality measure? J Neurosurg Pediatr 17:249–259
Simon TD, Hall M, Riva-Cambrin J, Albert JE, Jeffries HE, Lafleur B, Dean JM, Kestle JR (2009) Infection rates following initial cerebrospinal fluid shunt placement across pediatric hospitals in the United States. Clinical article. J Neurosurg Pediatr 4:156–165
Simon TD, Kronman MP, Whitlock KB, Gove N, Browd SR, Holubkov R, Kestle JR, Kulkarni AV, Langley M, Limbrick DD Jr, Luerssen TG, Oakes J, Riva-Cambrin J, Rozzelle C, Shannon C, Tamber M, Wellons JC 3rd, Whitehead WE, Mayer-Hamblett N (2016) Variability in management of first cerebrospinal fluid shunt infection: a prospective multi-institutional observational cohort study. J Pediatr 179:185–191.e182
Simon TD, Riva-Cambrin J, Srivastava R, Bratton SL, Dean JM, Kestle JR (2008) Hospital care for children with hydrocephalus in the United States: utilization, charges, comorbidities, and deaths. J Neurosurg Pediatr 1:131–137
Spader HS, Hertzler DA, Kestle JR, Riva-Cambrin J (2015) Risk factors for infection and the effect of an institutional shunt protocol on the incidence of ventricular access device infections in preterm infants. J Neurosurg Pediatr 15:156–160
Whitehead WE, Riva-Cambrin J, Kulkarni AV, Wellons JC 3rd, Rozzelle CJ, Tamber MS, Limbrick DD Jr, Browd SR, Naftel RP, Shannon CN, Simon TD, Holubkov R, Illner A, Cochrane DD, Drake JM, Luerssen TG, Oakes WJ, Kestle JR (2017) Ventricular catheter entry site and not catheter tip location predicts shunt survival: a secondary analysis of 3 large pediatric hydrocephalus studies. J Neurosurg Pediatr 19:157–167
Zou J, Han Y, So SS (2008) Overview of artificial neural networks. Methods Mol Biol 458:15–23
Kousi M, Katsanis N (2016) The genetic basis of hydrocephalus. Annu Rev Neurosci 39:409–435
Hale AT, Wellons JC, Limbrick DD, Schiff SJ, Gamazon ER (2020) Alterations in white matter and total brain volumes underlie genetic risk of hydrocephalus. Neurosurgery 67
Karimy JK, Zhang J, Kurland DB, Theriault BC, Duran D, Stokum JA, Furey CG, Zhou X, Mansuri MS, Montejo J, Vera A, DiLuna ML, Delpire E, Alper SL, Gunel M, Gerzanich V, Medzhitov R, Simard JM, Kahle KT (2017) Inflammation-dependent cerebrospinal fluid hypersecretion by the choroid plexus epithelium in posthemorrhagic hydrocephalus. Nat Med 23:997–1003
Sullivan W, Reeves BC, Duy PQ, Nelson-Williams C, Dong W, Jin SC, Kahle KT (2020) Exome sequencing as a potential diagnostic adjunct in sporadic congenital hydrocephalus. JAMA Pediatr
Brookes E, Shi Y (2014) Diverse epigenetic mechanisms of human disease. Annu Rev Genet 48:237–268
Iakoucheva LM, Muotri AR, Sebat J (2019) Getting to the cores of autism. Cell 178:1287–1298
Leu C, Stevelink R, Smith AW, Goleva SB, Kanai M, Ferguson L, Campbell C, Kamatani Y, Okada Y, Sisodiya SM, Cavalleri GL, Koeleman BPC, Lerche H, Jehi L, Davis LK, Najm IM, Palotie A, Daly MJ, Busch RM, Lal D (2019) Polygenic burden in focal and generalized epilepsies. Brain 142:3473–3481
Peck G, Smeeth L, Whittaker J, Casas JP, Hingorani A, Sharma P (2008) The genetics of primary haemorrhagic stroke, subarachnoid haemorrhage and ruptured intracranial aneurysms in adults. PLoS One 3:e3691
Southerland AM, Meschia JF, Worrall BB (2013) Shared associations of nonatherosclerotic, large-vessel, cerebrovascular arteriopathies: considering intracranial aneurysms, cervical artery dissection, moyamoya disease and fibromuscular dysplasia. Curr Opin Neurol 26:13–28
Rudin C (2019) Stop explaining black box machine learning models for high stakes decisions and use interpretable models instead. Nat Mach Intell 1:206–215
Bydon M, Schirmer CM, Oermann EK, Kitagawa RS, Pouratian N, Davies J, Sharan A, Chambless LB (2020) Big data defined: a practical review for neurosurgeons. World Neurosurg 133:e842–e849
Oravec CS, Motiwala M, Reed K, Jones TL, Klimo P Jr (2019) Big data research in pediatric neurosurgery: content, statistical output, and bibliometric analysis. Pediatr Neurosurg 54:85–97
Pasini A (2015) Artificial neural networks for small dataset analysis. J Thorac Dis 7:953–960
Hale AT, Stanton AN, Zhao S, Haji F, Gannon SR, Arynchyna A, Wellons JC, Rocque BG, Naftel RP (2019) Predictors of endoscopic third ventriculostomy ostomy status in patients who experience failure of endoscopic third ventriculostomy with choroid plexus cauterization. J Neurosurg Pediatr:1–6
Kulkarni AV, Riva-Cambrin J, Browd SR, Drake JM, Holubkov R, Kestle JR, Limbrick DD, Rozzelle CJ, Simon TD, Tamber MS, Wellons JC 3rd, Whitehead WE (2014) Endoscopic third ventriculostomy and choroid plexus cauterization in infants with hydrocephalus: a retrospective Hydrocephalus Clinical Research Network study. J Neurosurg Pediatr 14:224–229
Kulkarni AV, Riva-Cambrin J, Holubkov R, Browd SR, Cochrane DD, Drake JM, Limbrick DD, Rozzelle CJ, Simon TD, Tamber MS, Wellons JC 3rd, Whitehead WE, Kestle JR (2016) Endoscopic third ventriculostomy in children: prospective, multicenter results from the Hydrocephalus Clinical Research Network. J Neurosurg Pediatr 18:423–429
Kulkarni AV, Riva-Cambrin J, Rozzelle CJ, Naftel RP, Alvey JS, Reeder RW, Holubkov R, Browd SR, Cochrane DD, Limbrick DD, Simon TD, Tamber M, Wellons JC, Whitehead WE, Kestle JRW (2018) Endoscopic third ventriculostomy and choroid plexus cauterization in infant hydrocephalus: a prospective study by the Hydrocephalus Clinical Research Network. J Neurosurg Pediatr 21:214–223
Riva-Cambrin J, Kestle JRW, Rozzelle CJ, Naftel RP, Alvey JS, Reeder RW, Holubkov R, Browd SR, Cochrane DD, Limbrick DD, Shannon CN, Simon TD, Tamber MS, Wellons JC, Whitehead WE, Kulkarni AV (2019) Predictors of success for combined endoscopic third ventriculostomy and choroid plexus cauterization in a North American setting: a Hydrocephalus Clinical Research Network study. J Neurosurg Pediatr:1–11
Acknowledgements
The authors would like to thank their colleagues for their past and ongoing support of HCRN:
D Brockmeyer, M Walker, R Bollo, S Cheshier, J Blount, J Johnston, B Rocque, L Acakpo-Satchivi, WJ Oakes, J Rutka, M Taylor, P Dirks, D Curry, G Aldave, R Dauser, A Jea, S Lam, H Weiner, T Luerssen, R Ellenbogen, J Ojemann, A Lee, A Avellino, S Greene, E Tyler-Kabara, T Abel, TS Park, J Strahle, S McEvoy, M Smyth, N Tulipan, A Singhal, P Steinbok, D Cochrane, W Hader, C Gallagher, M Benour, E Kiehna, JG McComb, P Chiarelli, A Robison, A Alexander, M Handler, B O’Neill, C Wilkinson, L Governale, A Drapeau, J Leonard, E Sribnick, A Shaikhouni, E Ahn, A Cohen, M Groves, S Robinson, and CM Bonfield.
In addition, our work would not be possible without the outstanding support of the dedicated personnel at each clinical site and the data coordinating center. Special thanks go to the following: L Holman, J Clawson, P Martello, N Tattersall, T Bach (Salt Lake City); A Arynchyna, A Bey (Birmingham); H Ashrafpour, M Lamberti-Pasculli, L O’Connor (Toronto); E Sanchez, E Santisbon, S Martinez, S Ryan (Houston); A Anderson, G Bowen (Seattle); K Diamond, A Luther (Pittsburgh); A Morgan, H Botteron, D Morales, M Gabir, D Berger, D Mercer (St. Louis); A Wiseman, J Stoll, D Dawson, S Gannon (Nashville); A Cheong, R Hengel (British Columbia); R Rashid, S Ahmed (Calgary); M Alrefaie, R Daniel, A Loudermilk (Baltimore); N Rea, C Cook (Los Angeles); S Staulcup (Colorado); A Sheline (Columbus); and N Nunn, M Langley, V Wall, D Austin, B Conley, V Freimann, L Herrera, B Miller (Utah Data Coordinating Center).
Funding
The HCRN has been funded by National Institute of Neurological Disorders and Stroke (NINDS grant nos. 1U01NS107486-01A1 and 1RC1NS068943-01), Patient Centered Outcome Research Institute (PCORI grant no. CER-1403-13857), The Gerber Foundation (reference no. 1692-3638), private philanthropy, and the Hydrocephalus Association. A.T.H. is supported by the National Institutes of Health (F30HL143826) and Vanderbilt University Medical Scientist Training Program (5T32GM007347). None of the sponsors participated in design and conduct of this study; collection, management, analysis, and interpretation of the data; or preparation, review, or approval of this manuscript. Its contents are solely the responsibility of the authors and do not necessarily represent the official view of the sponsors.
Author information
Authors and Affiliations
Consortia
Corresponding author
Ethics declarations
Conflict of interest
None.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Previously presented as a platform talk at the AANS/CNS Pediatric Section Annual Meeting, Scottsdale AZ, December 8th,2019.
Appendix. Hydrocephalus Clinical Research Network
Appendix. Hydrocephalus Clinical Research Network
Members
The HCRN currently consists of the following clinical centers and investigators: Primary Children’s Hospital, University of Utah (J Kestle); Children’s Hospital of Alabama, University of Alabama at Birmingham (C Rozzelle); Hospital for Sick Children, University of Toronto (J Drake, A Kulkarni); Texas Children’s Hospital, Baylor College of Medicine (W Whitehead); Seattle Children’s Hospital, University of Washington (S Browd, T Simon, J Hauptman); Children’s Hospital of Pittsburgh, University of Pittsburgh (I Pollack); St. Louis Children’s Hospital, Washington University in St. Louis (D Limbrick); Monroe Carell Jr. Children’s Hospital at Vanderbilt, Vanderbilt University Medical Center (J Wellons, R Naftel, C Shannon); British Columbia Children’s Hospital, University of British Columbia (M Tamber, P McDonald); Alberta Children’s Hospital, University of Calgary (J Riva-Cambrin); The Johns Hopkins Hospital (E Jackson); Children’s Hospital of Los Angeles (M Krieger, J Chu); Children’s Hospital Colorado (T Hankinson); Nationwide Children’s Hospital (J Pindrik); HCRN Data Coordinating Center, Department of Pediatrics, University of Utah (R Holubkov).
Rights and permissions
About this article
Cite this article
Hale, A.T., Riva-Cambrin, J., Wellons, J.C. et al. Machine learning predicts risk of cerebrospinal fluid shunt failure in children: a study from the hydrocephalus clinical research network. Childs Nerv Syst 37, 1485–1494 (2021). https://doi.org/10.1007/s00381-021-05061-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00381-021-05061-7