Abstract
Objective
To investigate the efficacy of contrast-enhanced computed tomography (CECT)–based radiomics signatures for preoperative prediction of pathological grades of hepatocellular carcinoma (HCC) via machine learning.
Methods
In this single-center retrospective study, data collected from 297 consecutive subjects with HCC were allocated to training dataset (n = 237) and test dataset (n = 60). Manual segmentation of lesion sites was performed with ITK-SNAP, the radiomics features were extracted by the Pyradiomics, and radiomics signatures were synthesized using recursive feature elimination (RFE) method. The prediction models for pathological grading of HCC were established by using eXtreme Gradient Boosting (XGBoost). The performance of the models was evaluated using the AUC along with 95% confidence intervals (CIs) and standard deviation, sensitivity, specificity, and accuracy.
Results
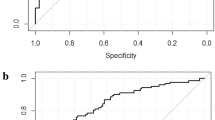
The radiomics signatures were found highly efficient for machine learning to differentiate high-grade HCC from low-grade HCC. For the clinical factors, when they were merely applied to train a machine learning model, the model achieved an AUC of 0.6698, along with 95% CI and standard deviation of 0.5307–0.8089 and 0.0710, respectively (sensitivity, 0.6522; specificity, 0.4595; accuracy, 0.5333). Meanwhile, when the radiomics signatures were applied in association with clinical factors to train a machine learning model, the performance of the model remarkably increased with AUC of 0.8014, along with 95% CI and standard deviation of 0.6899–0.9129 and 0.0569, respectively (sensitivity, 0.6522; specificity, 0.7297; accuracy, 0.7000).
Conclusions
The radiomics signatures could non-invasively explore the underlying association between CECT images and pathological grades of HCC.
Key Points
• The radiomics signatures may non-invasively explore the underlying association between CECT images and pathological grades of HCC via machine learning.
• The radiomics signatures of CECT images may enhance the prediction performance of pathological grading of HCC, and further validation is required.
• The features extracted from arterial phase CECT images may be more reliable than venous phase CECT images for predicting pathological grades of HCC.




Similar content being viewed by others
Abbreviations
- 3D:
-
Three-dimensional
- AFP:
-
Alpha-fetoprotein
- ALT:
-
Alanine aminotransferase
- AP:
-
Arterial phase
- AST:
-
Aspartate aminotransferase
- cc-RCC:
-
Clear cell renal carcinoma
- CECT:
-
Contrast-enhanced computed tomography
- CI:
-
Confidence interval
- CLAIM:
-
Checklist for AI in Medical Imaging
- DICOM:
-
Digital imaging and communications in medicine
- EHR:
-
Electronic health records
- ES:
-
Edmondson-Steiner
- GGT:
-
Gamma-glutamyl transpeptidase
- GLCM:
-
Gray-level co-occurrence matrix
- GLDM:
-
Gray-level dependence matrix
- GLRLM:
-
Gray-level run length matrix
- GLSZM:
-
Gray-level size zone matrix
- HBsAg:
-
Hepatitis B surface antigen
- HCVab:
-
Hepatitis C antibody
- ICC:
-
Intraclass correlation coefficient
- LT:
-
Liver transplantation
- MRI:
-
Magnetic resonance imaging
- MWA:
-
Microwave ablation
- NGTDM:
-
Neighboring gray tone difference matrix
- RFA:
-
Radiofrequency ablation
- RFE:
-
Recursive feature elimination
- ROI:
-
Region of interest
- SVM:
-
Support vector machine
- VP:
-
Venous phase
- XGBoost:
-
eXtreme Gradient Boosting
References
European Association for the Study of the Liver (2018) EASL clinical practice guidelines: management of hepatocellular carcinoma. J. Hepatol. 69(1):182–236
Njei B, Rotman Y, Ditah I, Lim JK (2015) Emerging trends in hepatocellular carcinoma incidence and mortality. Hepatology 61(1):191–199
Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A (2018) Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 68(6):394–424
Lencioni R, Crocetti L (2012) Local-regional treatment of hepatocellular carcinoma. Radiology 262(1):43–58
Forner A, Reig M, Bruix J (2018) Hepatocellular carcinoma. Lancet 391(10127):1301–1314
Wang Y-Y, Zhong J-H, Su Z-Y et al (2016) Albumin-bilirubin versus Child-Pugh score as a predictor of outcome after liver resection for hepatocellular carcinoma. Br J Surg 103(6):725–734
Sasaki A, Kai S, Iwashita Y, Hirano S, Ohta M, Kitano S (2005) Microsatellite distribution and indication for locoregional therapy in small hepatocellular carcinoma. Cancer 103(2):299–306
Weiss LM, Medeiros LJ, Vickery AL (1989) Pathologic features of prognostic significance in adrenocortical carcinoma. Am J Surg Pathol 13(3):202–206
Martins-Filho SN, Paiva C, Azevedo RS, Alves VAF (2017) Histological grading of hepatocellular carcinoma-a systematic review of literature. Front Med (Lausanne) 4:193
Bruix J, Sherman M (2005) Management of hepatocellular carcinoma. Hepatology 42(5):1208–1236
Llovet JM, Zucman-Rossi J, Pikarsky E et al (2016) Hepatocellular carcinoma. Nat Rev Dis Primers 2:16018
Robert M, Sofair AN, Thomas A et al (2009) A comparison of hepatopathologists’ and community pathologists' review of liver biopsy specimens from patients with hepatitis C. Clin Gastroenterol Hepatol 7(3):335–338
Gillies RJ, Kinahan PE, Hricak H (2016) Radiomics: images are more than pictures, they are data. Radiology 278(2):563–577
Huang Y, Liu Z, He L et al (2016) Radiomics signature: a potential biomarker for the prediction of disease-free survival in early-stage (I or II) non-small cell lung cancer. Radiology 281(3):947–957
Ortiz-Ramón R, Larroza A, Ruiz-España S, Arana E, Moratal D (2018) Classifying brain metastases by their primary site of origin using a radiomics approach based on texture analysis: a feasibility study. Eur Radiol 28(11):4514–4523
Jin X, Zheng X, Chen D et al (2019) Prediction of response after chemoradiation for esophageal cancer using a combination of dosimetry and CT radiomics. Eur Radiol 29(11):6080–6088
Hennedige T, Venkatesh SK (2013) Imaging of hepatocellular carcinoma: diagnosis, staging and treatment monitoring. Cancer Imaging 12:530–547
Oh J, Lee JM, Park J et al (2019) Hepatocellular carcinoma: texture analysis of preoperative computed tomography images can provide markers of tumor grade and disease-free survival. Korean J Radiol 20(4):569–579
Wu M, Tan H, Gao F et al (2019) Predicting the grade of hepatocellular carcinoma based on non-contrast-enhanced MRI radiomics signature. Eur Radiol 29(6):2802–2811
Willemink MJ, Koszek WA, Hardell C et al (2020) Preparing medical imaging data for machine learning. Radiology 295(1):4–15
Edmondson HA, Steiner PE (1954) Primary carcinoma of the liver: a study of 100 cases among 48,900 necropsies. Cancer 7(3):462–503
Yushkevich PA, Piven J, Hazlett HC et al (2006) User-guided 3D active contour segmentation of anatomical structures: significantly improved efficiency and reliability. Neuroimage 31(3):1116–1128
van Griethuysen JJM, Fedorov A, Parmar C et al (2017) Computational radiomics system to decode the radiographic phenotype. Cancer Res. 77(21):e104–e107
Leijenaar RTH, Nalbantov G, Carvalho S et al (2015) The effect of SUV discretization in quantitative FDG-PET radiomics: the need for standardized methodology in tumor texture analysis. Sci Rep 5:11075
E L, Lu L, Li L, Yang H, Schwartz LH, Zhao B (2019) Radiomics for classification of lung cancer histological subtypes based on nonenhanced computed tomography. Acad Radiol 26(9):1245–1252
Guyon I, Weston J, Barnhill S, Vapnik V (2002) Gene Selection for Cancer Classification using Support Vector Machines. Mach Learn 46(1/3):389–422
Koo TK, Li MY (2016) A guideline of selecting and reporting intraclass correlation coefficients for reliability research. J Chiropr Med 15(2):155–163
Leijenaar RTH, Carvalho S, Velazquez ER et al (2013) Stability of FDG-PET Radiomics features: an integrated analysis of test-retest and inter-observer variability. Acta Oncol. 52(7):1391–1397
Bektas CT, Kocak B, Yardimci AH et al (2019) Clear cell renal cell carcinoma: machine learning-based quantitative computed tomography texture analysis for prediction of Fuhrman nuclear grade. Eur Radiol 29(3):1153–1163
Singh A (2019) Foundations of machine learning. SSRN J. https://doi.org/10.2139/ssrn.3399990
Chen T, Guestrin C (2016) XGBoost: a scalable tree boosting system. In: Krishnapuram B, Shah M, Smola A, Aggarwal C, Shen D, Rastogi R (eds) Proceedings of the 22nd ACM SIGKDD international conference on knowledge discovery and data mining. ACM, New York, pp 785–794
Akinkunmi M (2019) Introduction to statistics using R. Morgan & Claypool, San Rafael California
Mongan J, Moy L, Kahn CE (2020) Checklist for artificial intelligence in medical imaging (CLAIM): a guide for authors and reviewers. Radiol Artif Intell 2(2):e200029
Zhou W, Zhang L, Wang K et al (2017) Malignancy characterization of hepatocellular carcinomas based on texture analysis of contrast-enhanced MR images. J Magn Reson Imaging 45(5):1476–1484
Parmar C, Grossmann P, Bussink J, Lambin P, Aerts HJWL (2015) Machine learning methods for quantitative radiomic biomarkers. Sci Rep 5:13087
Sun P, Wang D, Mok VC, Shi L (2019) Comparison of feature selection methods and machine learning classifiers for radiomics analysis in glioma grading. IEEE Access 7:102010–102020
Funding
The study was funded by the National Natural Science Foundation of China (NO. 71974065) and Key R & D and promotion projects in Henan Province (NO. 182400410172).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Guarantor
The scientific guarantor of this publication is Jingdong Ma.
Conflict of interest
The authors of this manuscript declare no relationships with any companies, whose products or services may be related to the subject matter of the article.
Statistics and biometry
No complex statistical methods were necessary for this paper.
Informed consent
Written informed consent was obtained from all subjects (patients) in this study.
Ethical approval
Institutional Review Board approval was obtained.
Methodology
• Retrospective
• Diagnostic or prognostic study
• Performed at one institution
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
ESM 1
(PNG 493 kb)
ESM 2
(PNG 1033 kb)
ESM 3
(PNG 794 kb)
ESM 4
(PNG 1026 kb)
ESM 5
(PNG 718 kb)
ESM 6
(PNG 1026 kb)
ESM 7
(PNG 1537 kb)
ESM 8
Details of the various features and filters (DOCX 13 kb)
ESM 9
The Checklist for AI in Medical Imaging (CLAIM) (DOCX 24 kb)
ESM 10
The list of the extracted features in our study (CSV 127 kb)
ESM 11
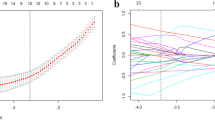
The results of feature selection and feature importance. The results of feature selection of AP images, VP images, clinical factors, AP-VP images, combined clinical factors and AP images, combined clinical factors and VP images, combined clinical factors and AP-VP images are shown in an order from A to G, respectively.5.The ROC curves with confidence intervalof AP images, VP images, clinical factors, AP-VP images, combined clinical factors and AP images, combined clinical factors and VP images are shown in an order from A to F, respectively. (PNG 6385 kb)
Rights and permissions
About this article
Cite this article
Mao, B., Zhang, L., Ning, P. et al. Preoperative prediction for pathological grade of hepatocellular carcinoma via machine learning–based radiomics. Eur Radiol 30, 6924–6932 (2020). https://doi.org/10.1007/s00330-020-07056-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00330-020-07056-5