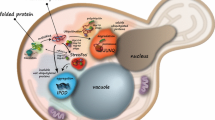
Abstract
Coalescence of proteins into different types of intracellular bodies has surfaced as a widespread adaptive mechanism to re-organize cells and cellular functions in response to specific cues. These structures, composed of proteins or protein-mRNA-complexes, regulate cellular processes through modulating enzymatic activities, gene expression or shielding macromolecules from damage. Accordingly, such bodies are associated with a wide-range of processes, including meiosis, memory-encoding, host-pathogen interactions, cancer, stress responses, as well as protein quality control, DNA replication stress and aneuploidy. Importantly, these distinct coalescence responses are controlled, and in many cases regulated by chaperone proteins. While cells can tolerate and proficiently coordinate numerous distinct types of protein bodies, some of them are also intimately linked to diseases or the adverse effects of aging. Several protein bodies that differ in composition, packing, dynamics, size, and localization were originally discovered in budding yeast. Here, we provide a concise and comparative review of their nature and nomenclature.

Similar content being viewed by others
References
Aguilaniu H, Gustafsson L, Rigoulet M, Nystrom T (2003) Asymmetric inheritance of oxidatively damaged proteins during cytokinesis. Science 299:1751–1753. doi:10.1126/science.1080418
Alberti S, Halfmann R, King O, Kapila A, Lindquist S (2009) A systematic survey identifies prions and illuminates sequence features of prionogenic proteins. Cell 137:146–158. doi:10.1016/j.cell.2009.02.044
Allen C, Buttner S, Aragon AD, Thomas JA, Meirelles O, Jaetao JE, Benn D, Ruby SW, Veenhuis M, Madeo F, Werner-Washburne M (2006) Isolation of quiescent and nonquiescent cells from yeast stationary-phase cultures. J Cell Biol 174:89–100. doi:10.1083/jcb.200604072
Andersson V, Hanzen S, Liu B, Molin M, Nystrom T (2013) Enhancing protein disaggregation restores proteasome activity in aged cells. Aging 5:802–812
Berchowitz LE, Kabachinski G, Walker MR, Carlile TM, Gilbert WV, Schwartz TU, Amon A (2015) Regulated formation of an amyloid-like translational repressor governs gametogenesis. Cell 163:406–418. doi:10.1016/j.cell.2015.08.060
Breker M, Gymrek M, Moldavski O, Schuldiner M (2014) LoQAtE–Localization and Quantitation ATlas of the yeast proteomE: a new tool for multiparametric dissection of single-protein behavior in response to biological perturbations in yeast. Nucl Acid Res 42:D726–D730. doi:10.1093/nar/gkt933
Buchan JR, Muhlrad D, Parker R (2008) P bodies promote stress granule assembly in Saccharomyces cerevisiae. J Cell Biol 183:441–455. doi:10.1083/jcb.200807043
Buchan JR, Yoon JH, Parker R (2011) Stress-specific composition, assembly and kinetics of stress granules in Saccharomyces cerevisiae. J Cell Sci 124:228–239. doi:10.1242/jcs.078444
Carroll JS, Munchel SE, Weis K (2011) The DExD/H box ATPase Dhh1 functions in translational repression, mRNA decay, and processing body dynamics. J Cell Biol 194:527–537. doi:10.1083/jcb.201007151
Castello A, Fischer B, Eichelbaum K, Horos R, Beckmann BM, Strein C, Davey NE, Humphreys DT, Preiss T, Steinmetz LM, Krijgsveld J, Hentze MW (2012) Insights into RNA biology from an atlas of mammalian mRNA-binding proteins. Cell 149:1393–1406. doi:10.1016/j.cell.2012.04.031
Caudron F, Barral Y (2013) A super-assembly of Whi3 encodes memory of deceptive encounters by single cells during yeast courtship. Cell 155:1244–1257. doi:10.1016/j.cell.2013.10.046
Cherkasov V, Hofmann S, Druffel-Augustin S, Mogk A, Tyedmers J, Stoecklin G, Bukau B (2013) Coordination of translational control and protein homeostasis during severe heat stress. Curr Biol CB 23:2452–2462. doi:10.1016/j.cub.2013.09.058
Clay L, Caudron F, Denoth-Lippuner A, Boettcher B, Buvelot Frei S, Snapp EL, Barral Y (2014) A sphingolipid-dependent diffusion barrier confines ER stress to the yeast mother cell. eLife 3:e01883. doi:10.7554/eLife.01883
Coelho M, Lade SJ, Alberti S, Gross T, Tolic IM (2014) Fusion of protein aggregates facilitates asymmetric damage segregation. PLoS Biol 12:e1001886. doi:10.1371/journal.pbio.1001886
De Virgilio C (2012) The essence of yeast quiescence. FEMS Microbiol Rev 36:306–339. doi:10.1111/j.1574-6976.2011.00287.x
Decker CJ, Parker R (2012) P-bodies and stress granules: possible roles in the control of translation and mRNA degradation. Cold Spring Harb Perspect Biol 4:a012286. doi:10.1101/cshperspect.a012286
Decker CJ, Teixeira D, Parker R (2007) Edc3p and a glutamine/asparagine-rich domain of Lsm4p function in processing body assembly in Saccharomyces cerevisiae. J Cell Biol 179:437–449. doi:10.1083/jcb.200704147
Denoth Lippuner A, Julou T, Barral Y (2014) Budding yeast as a model organism to study the effects of age. FEMS Microbiol Rev 38:300–325. doi:10.1111/1574-6976.12060
Denoth-Lippuner A, Krzyzanowski MK, Stober C, Barral Y (2014) Role of SAGA in the asymmetric segregation of DNA circles during yeast ageing. eLife 3:e03790. doi:10.7554/eLife.03790
Doronina VA, Staniforth GL, Speldewinde SH, Tuite MF, Grant CM (2015) Oxidative stress conditions increase the frequency of de novo formation of the yeast [PSI+] prion. Mol Microbiol 96:163–174. doi:10.1111/mmi.12930
Duennwald ML (2013) Yeast as a platform to explore polyglutamine toxicity and aggregation. Methods Mol Biol 1017:153–161. doi:10.1007/978-1-62703-438-8_11
Duennwald ML, Echeverria A, Shorter J (2012) Small heat shock proteins potentiate amyloid dissolution by protein disaggregases from yeast and humans. PLoS Biol 10:e1001346. doi:10.1371/journal.pbio.1001346
Elden AC, Kim HJ, Hart MP, Chen-Plotkin AS, Johnson BS, Fang X, Armakola M, Geser F, Greene R, Lu MM, Padmanabhan A, Clay-Falcone D, McCluskey L, Elman L, Juhr D, Gruber PJ, Rub U, Auburger G, Trojanowski JQ, Lee VM, Van Deerlin VM, Bonini NM, Gitler AD (2010) Ataxin-2 intermediate-length polyglutamine expansions are associated with increased risk for ALS. Nature 466:1069–1075. doi:10.1038/nature09320
Erjavec N, Larsson L, Grantham J, Nystrom T (2007) Accelerated aging and failure to segregate damaged proteins in Sir2 mutants can be suppressed by overproducing the protein aggregation-remodeling factor Hsp104p. Genes Dev 21:2410–2421. doi:10.1101/gad.439307
Escusa-Toret S, Vonk WI, Frydman J (2013) Spatial sequestration of misfolded proteins by a dynamic chaperone pathway enhances cellular fitness during stress. Nat Cell Biol 15:1231–1243. doi:10.1038/ncb2838
Fang NN, Ng AH, Measday V, Mayor T (2011) Hul5 HECT ubiquitin ligase plays a major role in the ubiquitylation and turnover of cytosolic misfolded proteins. Nat Cell Biol 13:1344–1352. doi:10.1038/ncb2343
Fang NN, Chan GT, Zhu M, Comyn SA, Persaud A, Deshaies RJ, Rotin D, Gsponer J, Mayor T (2014) Rsp5/Nedd4 is the main ubiquitin ligase that targets cytosolic misfolded proteins following heat stress. Nat Cell Biol 16:1227–1237. doi:10.1038/ncb3054
Fioriti L, Myers C, Huang YY, Li X, Stephan JS, Trifilieff P, Colnaghi L, Kosmidis S, Drisaldi B, Pavlopoulos E, Kandel ER (2015) The persistence of hippocampal-based memory requires protein synthesis mediated by the prion-like protein CPEB3. Neuron 86:1433–1448. doi:10.1016/j.neuron.2015.05.021
Garcia DM, Jarosz DF (2014) Rebels with a cause: molecular features and physiological consequences of yeast prions. FEMS Yeast Res 14:136–147
Herrmann L, Wiegmann C, Arsalan-Werner A, Hilbrich I, Jager C, Flach K, Suttkus A, Lachmann I, Arendt T, Holzer M (2015) Hook proteins: association with Alzheimer pathology and regulatory role of hook3 in amyloid beta generation. PLoS One 10:e0119423. doi:10.1371/journal.pone.0119423
Hill SM, Hao X, Liu B, Nystrom T (2014) Life-span extension by a metacaspase in the yeast Saccharomyces cerevisiae. Science 344:1389–1392. doi:10.1126/science.1252634
Ho YH, Gash AP (2015) Exploiting the yeast stress-activated signaling network to inform on stress biology and disease signaling. Curr Genet 61:503–511
Holmes WM, Klaips CL, Serio TR (2014) Defining the limits: protein aggregation and toxicity in vivo. Crit Rev Biochem Mol Biol 49:294–303. doi:10.3109/10409238.2014.914151
Hoyle NP, Castelli LM, Campbell SG, Holmes LE, Ashe MP (2007) Stress-dependent relocalization of translationally primed mRNPs to cytoplasmic granules that are kinetically and spatially distinct from P-bodies. J Cell Biol 179:65–74. doi:10.1083/jcb.200707010
Jain S, Wheeler JR, Walters RW, Agrawal A, Barsic A, Parker R (2016) ATPase-modulated stress granules contain a diverse proteome and substructure. Cell 164:487–498. doi:10.1016/j.cell.2015.12.038
Janssens GE, Meinema AC, Gonzalez J, Wolters JC, Schmidt A, Guryev V, Bischoff R, Wit EC, Veenhoff LM, Heinemann M (2015) Protein biogenesis machinery is a driver of replicative aging in yeast. eLife 4:e08527. doi:10.7554/eLife.08527
Kaganovich D, Kopito R, Frydman J (2008) Misfolded proteins partition between two distinct quality control compartments. Nature 454:1088–1095. doi:10.1038/nature07195
Kayatekin C, Matlack KE, Hesse WR, Guan Y, Chakrabortee S, Russ J, Wanker EE, Shah JV, Lindquist S (2014) Prion-like proteins sequester and suppress the toxicity of huntingtin exon 1. Proc Natl Acad Sci USA 111:12085–12090. doi:10.1073/pnas.1412504111
Khurana V, Tardiff DF, Chung CY, Lindquist S (2015) Toward stem cell-based phenotypic screens for neurodegenerative diseases. Nat Rev Neurol 11:339–350. doi:10.1038/nrneurol.2015.79
Kim HJ, Kim NC, Wang YD, Scarborough EA, Moore J, Diaz Z, MacLea KS, Freibaum B, Li S, Molliex A, Kanagaraj AP, Carter R, Boylan KB, Wojtas AM, Rademakers R, Pinkus JL, Greenberg SA, Trojanowski JQ, Traynor BJ, Smith BN, Topp S, Gkazi AS, Miller J, Shaw CE, Kottlors M, Kirschner J, Pestronk A, Li YR, Ford AF, Gitler AD, Benatar M, King OD, Kimonis VE, Ross ED, Weihl CC, Shorter J, Taylor JP (2013) Mutations in prion-like domains in hnRNPA2B1 and hnRNPA1 cause multisystem proteinopathy and ALS. Nature 495:467–473. doi:10.1038/nature11922
King OD, Gitler AD, Shorter J (2012) The tip of the iceberg: RNA-binding proteins with prion-like domains in neurodegenerative disease. Brain Res 1462:61–80. doi:10.1016/j.brainres.2012.01.016
Kroschwald S, Maharana S, Mateju D, Malinovska L, Nuske E, Poser I, Richter D, Alberti S (2015) Promiscuous interactions and protein disaggregases determine the material state of stress-inducible RNP granules. eLife 4:e06807. doi:10.7554/eLife.06807
Laporte D, Salin B, Daignan-Fornier B, Sagot I (2008) Reversible cytoplasmic localization of the proteasome in quiescent yeast cells. J Cell Biol 181:737–745. doi:10.1083/jcb.200711154
Li P, Banjade S, Cheng HC, Kim S, Chen B, Guo L, Llaguno M, Hollingsworth JV, King DS, Banani SF, Russo PS, Jiang QX, Nixon BT, Rosen MK (2012) Phase transitions in the assembly of multivalent signalling proteins. Nature 483:336–340. doi:10.1038/nature10879
Li YR, King OD, Shorter J, Gitler AD (2013) Stress granules as crucibles of ALS pathogenesis. J Cell Biol 201:361–372. doi:10.1083/jcb.201302044
Liebman SW, Chernoff YO (2012) Prions in yeast. Genetics 191:1041–1072. doi:10.1534/genetics.111.137760
Liu B, Larsson L, Caballero A, Hao X, Oling D, Grantham J, Nystrom T (2010) The polarisome is required for segregation and retrograde transport of protein aggregates. Cell 140:257–267. doi:10.1016/j.cell.2009.12.031
Liu IC, Chiu SW, Lee HY, Leu JY (2012) The histone deacetylase Hos2 forms an Hsp42-dependent cytoplasmic granule in quiescent yeast cells. Mol Biol Cell 23:1231–1242. doi:10.1091/mbc.E11-09-0752
Longo VD, Shadel GS, Kaeberlein M, Kennedy B (2012) Replicative and chronological aging in Saccharomyces cerevisiae. Cell Metab 16:18–31. doi:10.1016/j.cmet.2012.06.002
Majumdar A, Cesario WC, White-Grindley E, Jiang H, Ren F, Khan MR, Li L, Choi EM, Kannan K, Guo F, Unruh J, Slaughter B, Si K (2012) Critical role of amyloid-like oligomers of Drosophila Orb2 in the persistence of memory. Cell 148:515–529. doi:10.1016/j.cell.2012.01.004
Malinovska L, Kroschwald S, Munder MC, Richter D, Alberti S (2012) Molecular chaperones and stress-inducible protein-sorting factors coordinate the spatiotemporal distribution of protein aggregates. Mol Biol Cell 23:3041–3056. doi:10.1091/mbc.E12-03-0194
Malinovska L, Kroschwald S, Alberti S (2013) Protein disorder, prion propensities, and self-organizing macromolecular collectives. Biochim Biophys Acta 1834:918–931. doi:10.1016/j.bbapap.2013.01.003
Miller SB, Ho CT, Winkler J, Khokhrina M, Neuner A, Mohamed MY, Guilbride DL, Richter K, Lisby M, Schiebel E, Mogk A, Bukau B (2015) Compartment-specific aggregases direct distinct nuclear and cytoplasmic aggregate deposition. EMBO J 34:778–797. doi:10.15252/embj.201489524
Moldavski O, Amen T, Levin-Zaidman S, Eisenstein M, Rogachev I, Brandis A, Kaganovich D, Schuldiner M (2015) Lipid droplets are essential for efficient clearance of cytosolic inclusion bodies. Dev Cell 33:603–610. doi:10.1016/j.devcel.2015.04.015
Narayanaswamy R, Levy M, Tsechansky M, Stovall GM, O’Connell JD, Mirrielees J, Ellington AD, Marcotte EM (2009) Widespread reorganization of metabolic enzymes into reversible assemblies upon nutrient starvation. Proc Natl Acad Sci USA 106:10147–10152. doi:10.1073/pnas.0812771106
Newby GA, Lindquist S (2013) Blessings in disguise: biological benefits of prion-like mechanisms. Trends Cell Biol 23:251–259. doi:10.1016/j.tcb.2013.01.007
Noree C, Sato BK, Broyer RM, Wilhelm JE (2010) Identification of novel filament-forming proteins in Saccharomyces cerevisiae and Drosophila melanogaster. J Cell Biol 190:541–551. doi:10.1083/jcb.201003001
Nostramo R, Herman PK (2016) Deubiquitination and the regulation of stress granule assembly. Curr Genet. doi:10.1007/s00294-016-0571-9
Nostramo R, Varia SN, Zhang B, Emerson MM, Herman PK (2015) The catalytic activity of the Ubp3 deubiquitinating protease is required for efficient stress granule assembly in Saccharomyces cerevisiae. Mol Cell Biol 36:173–183
Oling D, Eisele F, Kvint K, Nystrom T (2014) Opposing roles of Ubp3-dependent deubiquitination regulate replicative life span and heat resistance. EMBO J 33:747–761. doi:10.1002/embj.201386822
Orij R, Postmus J, Ter Beek A, Brul S, Smits GJ (2009) In vivo measurement of cytosolic and mitochondrial pH using a pH-sensitive GFP derivative in Saccharomyces cerevisiae reveals a relation between intracellular pH and growth. Microbiology 155:268–278. doi:10.1099/mic.0.022038-0
Oromendia AB, Dodgson SE, Amon A (2012) Aneuploidy causes proteotoxic stress in yeast. Genes Dev 26:2696–2708. doi:10.1101/gad.207407.112
Park SH, Kukushkin Y, Gupta R, Chen T, Konagai A, Hipp MS, Hayer-Hartl M, Hartl FU (2013) PolyQ proteins interfere with nuclear degradation of cytosolic proteins by sequestering the Sis1p chaperone. Cell 154:134–145. doi:10.1016/j.cell.2013.06.003
Parsell DA, Kowal AS, Singer MA, Lindquist S (1994) Protein disaggregation mediated by heat-shock protein Hsp104. Nature 372:475–478. doi:10.1038/372475a0
Peters LZ, Hazan R, Breker M, Schuldiner M, Ben-Aroya S (2013) Formation and dissociation of proteasome storage granules are regulated by cytosolic pH. J Cell Biol 201:663–671. doi:10.1083/jcb.201211146
Peters LZ, Karmon O, Miodownik S, Ben-Aroya S (2016) Proteasome storage granules are transiently associated with the insoluble protein deposit (IPOD). J cell sci. doi:10.1242/jcs.179648
Petrovska I, Nuske E, Munder MC, Kulasegaran G, Malinovska L, Kroschwald S, Richter D, Fahmy K, Gibson K, Verbavatz JM, Alberti S (2014) Filament formation by metabolic enzymes is a specific adaptation to an advanced state of cellular starvation. eLife 3:e02409. doi:10.7554/elife.02409
Prasad R, Kawaguchi S, Ng DT (2010) A nucleus-based quality control mechanism for cytosolic proteins. Mol Biol Cell 21:2117–2127. doi:10.1091/mbc.E10-02-0111
Ramachandran V, Shah KH, Herman PK (2011) The cAMP-dependent protein kinase signaling pathway is a key regulator of P body foci formation. Mol Cell 43:973–981. doi:10.1016/j.molcel.2011.06.032
Rampelt H, Kirstein-Miles J, Nillegoda NB, Chi K, Scholz SR, Morimoto RI, Bukau B (2012) Metazoan Hsp70 machines use Hsp110 to power protein disaggregation. EMBO J 31:4221–4235. doi:10.1038/emboj.2012.264
Reijns MA, Alexander RD, Spiller MP, Beggs JD (2008) A role for Q/N-rich aggregation-prone regions in P-body localization. J Cell Sci 121:2463–2472. doi:10.1242/jcs.024976
Saarikangas J, Barral Y (2015) Protein aggregates are associated with replicative aging without compromising protein quality control. eLife 4:e06197. doi: 10.7554/eLife.06197
Sagot I, Pinson B, Salin B, Daignan-Fornier B (2006) Actin bodies in yeast quiescent cells: an immediately available actin reserve? Mol Biol Cell 17:4645–4655. doi:10.1091/mbc.E06-04-0282
Shah KH, Zhang B, Ramachandran V, Herman PK (2013) Processing body and stress granule assembly occur by independent and differentially regulated pathways in Saccharomyces cerevisiae. Genetics 193:109–123. doi:10.1534/genetics.112.146993
Shah KH, Nostramo R, Zhang B, Varia SN, Klett BM, Herman PK (2014) Protein kinases are associated with multiple, distinct cytoplasmic granules in quiescent yeast cells. Genetics 198:1495–1512. doi:10.1534/genetics.114.172031
Shcheprova Z, Baldi S, Frei SB, Gonnet G, Barral Y (2008) A mechanism for asymmetric segregation of age during yeast budding. Nature 454:728–734. doi:10.1038/nature07212
Shorter J (2011) The mammalian disaggregase machinery: Hsp110 synergizes with Hsp70 and Hsp40 to catalyze protein disaggregation and reactivation in a cell-free system. PLoS One 6:e26319. doi:10.1371/journal.pone.0026319
Shorter J, Lindquist S (2004) Hsp104 catalyzes formation and elimination of self-replicating Sup35 prion conformers. Science 304:1793–1797. doi:10.1126/science.1098007
Si K (2015) Prions: what are they good for? Annu Rev Cell Dev Biol 31:149–169. doi:10.1146/annurev-cellbio-100913-013409
Sinclair DA, Guarente L (1997) Extrachromosomal rDNA circles: a cause of aging in yeast. Cell 91:1033–1042
Song J, Yang Q, Yang J, Larsson L, Hao X, Zhu X, Malmgren-Hill S, Cvijovic M, Fernandez-Rodriguez J, Grantham J, Gustafsson CM, Liu B, Nystrom T (2014) Essential genetic interactors of SIR2 required for spatial sequestration and asymmetrical inheritance of protein aggregates. PLoS Genet 10:e1004539. doi:10.1371/journal.pgen.1004539
Specht S, Miller SB, Mogk A, Bukau B (2011) Hsp42 is required for sequestration of protein aggregates into deposition sites in Saccharomyces cerevisiae. J Cell Biol 195:617–629. doi:10.1083/jcb.201106037
Spokoini R, Moldavski O, Nahmias Y, England JL, Schuldiner M, Kaganovich D (2012) Confinement to organelle-associated inclusion structures mediates asymmetric inheritance of aggregated protein in budding yeast. Cell reports 2:738–747 doi:10.1016/j.celrep.2012.08.024
Sun Z, Diaz Z, Fang X, Hart MP, Chesi A, Shorter J, Gitler AD (2011) Molecular determinants and genetic modifiers of aggregation and toxicity for the ALS disease protein FUS/TLS. PLoS Biol 9:e1000614. doi:10.1371/journal.pbio.1000614
Suresh HG, da Silveira Dos Santos AX, Kukulski W, Tyedmers J, Riezman H, Bukau B, Mogk A (2015) Prolonged starvation drives reversible sequestration of lipid biosynthetic enzymes and organelle reorganization in Saccharomyces cerevisiae. Mol Biol Cell 26:1601–1615. doi:10.1091/mbc.E14-11-1559
Szebenyi G, Wigley WC, Hall B, Didier A, Yu M, Thomas P, Kramer H (2007) Hook2 contributes to aggresome formation. BMC Cell Biol 8:19. doi:10.1186/1471-2121-8-19
Takahara T, Maeda T (2012) Transient sequestration of TORC1 into stress granules during heat stress. Mol Cell 47:242–252. doi:10.1016/j.molcel.2012.05.019
Teixeira D, Parker R (2007) Analysis of P-body assembly in Saccharomyces cerevisiae. Mol Biol Cell 18:2274–2287. doi:10.1091/mbc.E07-03-0199
Teixeira D, Sheth U, Valencia-Sanchez MA, Brengues M, Parker R (2005) Processing bodies require RNA for assembly and contain nontranslating mRNAs. RNA 11:371–382. doi:10.1261/rna.7258505
Thayer NH, Leverich CK, Fitzgibbon MP, Nelson ZW, Henderson KA, Gafken PR, Hsu JJ, Gottschling DE (2014) Identification of long-lived proteins retained in cells undergoing repeated asymmetric divisions. Proc Natl Acad Sci USA 111:14019–14026. doi:10.1073/pnas.1416079111
Tkach JM, Yimit A, Lee AY, Riffle M, Costanzo M, Jaschob D, Hendry JA, Ou J, Moffat J, Boone C, Davis TN, Nislow C, Brown GW (2012) Dissecting DNA damage response pathways by analysing protein localization and abundance changes during DNA replication stress. Nat Cell Biol 14:966–976. doi:10.1038/ncb2549
Tuite MF (2015) Yeast prions: paramutation at the protein level? Semin Cell Dev Biol 44:51–61. doi:10.1016/j.semcdb.2015.08.016
Tyedmers J, Treusch S, Dong J, McCaffery JM, Bevis B, Lindquist S (2010) Prion induction involves an ancient system for the sequestration of aggregated proteins and heritable changes in prion fragmentation. Proc Natl Acad Sci USA 107:8633–8638. doi:10.1073/pnas.1003895107
Unal E, Kinde B, Amon A (2011) Gametogenesis eliminates age-induced cellular damage and resets life span in yeast. Science 332:1554–1557. doi:10.1126/science.1204349
Vabulas RM, Raychaudhuri S, Hayer-Hartl M, Hartl FU (2010) Protein folding in the cytoplasm and the heat shock response. Cold Spring Harb Perspect Biol 2:a004390. doi:10.1101/cshperspect.a004390
Wallace EW, Kear-Scott JL, Pilipenko EV, Schwartz MH, Laskowski PR, Rojek AE, Katanski CD, Riback JA, Dion MF, Franks AM, Airoldi EM, Pan T, Budnik BA, Drummond DA (2015) Reversible, specific, active aggregates of endogenous proteins assemble upon heat stress. Cell 162:1286–1298. doi:10.1016/j.cell.2015.08.041
Wang JT, Smith J, Chen BC, Schmidt H, Rasoloson D, Paix A, Lambrus BG, Calidas D, Betzig E, Seydoux G (2014) Regulation of RNA granule dynamics by phosphorylation of serine-rich, intrinsically disordered proteins in C. elegans. Life 3:e04591. doi:10.7554/eLife.04591
Weber SC, Brangwynne CP (2012) Getting RNA and protein in phase. Cell 149:1188–1191. doi:10.1016/j.cell.2012.05.022
Weisberg SJ, Lyakhovetsky R, Werdiger AC, Gitler AD, Soen Y, Kaganovich D (2012) Compartmentalization of superoxide dismutase 1 (SOD1G93A) aggregates determines their toxicity. Proc Natl Acad Sci USA 109:15811–15816. doi:10.1073/pnas.1205829109
Wickner RB, Shewmaker FP, Bateman DA, Edskes HK, Gorkovskiy A, Dayani Y, Bezsonov EE (2015) Yeast prions: structure, biology, and prion-handling systems. Microbiol Mol Biol Rev 79:1–17. doi:10.1128/MMBR.00041-14
Woerner AC, Frottin F, Hornburg D, Feng LR, Meissner F, Patra M, Tatzelt J, Mann M, Winklhofer KF, Hartl FU, Hipp MS (2016) Cytoplasmic protein aggregates interfere with nucleocytoplasmic transport of protein and RNA. Science 351:173–176. doi:10.1126/science.aad2033
Wolfe KJ, Ren HY, Trepte P, Cyr DM (2013) The Hsp70/90 cochaperone, Sti1, suppresses proteotoxicity by regulating spatial quality control of amyloid-like proteins. Mol Biol Cell 24:3588–3602. doi:10.1091/mbc.E13-06-0315
Yamamoto Y, Izawa S (2013) Adaptive response in stress granule formation and bulk translational repression upon a combined stress of mild heat shock and mild ethanol stress in yeast. Genes Cells 18:974–984. doi:10.1111/gtc.12090
Yang J, McCormick MA, Zheng J, Xie Z, Tsuchiya M, Tsuchiyama S, El-Samad H, Ouyang Q, Kaeberlein M, Kennedy BK, Li H (2015) Systematic analysis of asymmetric partitioning of yeast proteome between mother and daughter cells reveals “aging factors” and mechanism of lifespan asymmetry. Proc Natl Acad Sci USA 112:11977–11982. doi:10.1073/pnas.1506054112
Zhang H, Elbaum-Garfinkle S, Langdon EM, Taylor N, Occhipinti P, Bridges AA, Brangwynne CP, Gladfelter AS (2015) RNA controls PolyQ protein phase transitions. Mol Cell 60:220–230. doi:10.1016/j.molcel.2015.09.017
Zhou C, Slaughter BD, Unruh JR, Eldakak A, Rubinstein B, Li R (2011) Motility and segregation of Hsp104-associated protein aggregates in budding yeast. Cell 147:1186–1196. doi:10.1016/j.cell.2011.11.002
Zhou C, Slaughter BD, Unruh JR, Guo F, Yu Z, Mickey K, Narkar A, Ross RT, McClain M, Li R (2014) Organelle-based aggregation and retention of damaged proteins in asymmetrically dividing cells. Cell 159:530–542 doi:10.1016/j.cell.2014.09.026
Acknowledgments
We apologize to colleagues whose work we were unable to cite due to length restrains. We are thankful to Fabrice Caudron, Marek Krzyzanowski and Asim Sengör for comments on the manuscript. JS acknowledges FEBS and the Finnish Cultural Foundation and YB the European Research council and the ETH Zurich for financial support.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Communicated by M. Kupiec.
Rights and permissions
About this article
Cite this article
Saarikangas, J., Barral, Y. Protein aggregation as a mechanism of adaptive cellular responses. Curr Genet 62, 711–724 (2016). https://doi.org/10.1007/s00294-016-0596-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00294-016-0596-0