Abstract
The spread of antibiotic resistance is rapidly threatening the effectiveness of antibiotics in the clinical setting. Many infections are being caused by known and unknown pathogenic bacteria that are resistant to many or all antibiotics currently available. Empedobacter falsenii is a nosocomial pathogen that can cause human infections. E. falsenii Wf282 strain was found to be resistant to many antibiotics, including carbapenems and colistin. Whole-genome shotgun sequencing of the strain was performed, and distinct features were identified. A novel metallo-β-lactamase, named EBR-2, was found, suggesting a potential role of E. falsenii as a reservoir of β-lactamases and other resistance determinants also found in its genome. The EBR-2 protein showed the highest catalytic efficiency for penicillin G as compared to meropenem and ampicillin and was unable to hydrolyze cefepime. The results described in this work broaden the current understanding of the role of β-lactamases in the Flavobacteriaceae family and suggest that E. falsenii Wf282 may be a reservoir of these novel resistance determinants.

Similar content being viewed by others

References
Barlam TF, Gupta K (2015) Antibiotic resistance spreads internationally across borders. J Law Med Ethics 43(Suppl 3):12–16
Anonymous (2014) Antimicrobial resistance: global Report on surveillance. World Health Organization
Anonymous (2013) Antibiotic resistance threats in the United States. Centers for Disease Control and Prevention (CDC)
Taconnelli E, Magrini N (2017) Global priority list of antibiotic-resistant bacteria to guide research, discovery, and development of new antibiotics, World Health Organization, Tübingen University, pp 1–7
Berrazeg M, Jeannot K, Ntsogo Enguéné VY, Broutin I, Loeffert S, Fournier D, Plésiat P (2015) Mutations in β-lactamase AmpC increase resistance of Pseudomonas aeruginosa isolates to antipseudomonal cephalosporins. Antimicrob Agents Chemother 59:6248–6255
Bhattacharya S, Bir R, Majumdar T (2015) Evaluation of multidrug resistant Staphylococcus aureus and their association with biofilm production in a Tertiary Care Hospital, Tripura, Northeast India. J Clin Diagn Res 9:DC01
Michiels B, Appelen L, Franck B, den Heijer CD, Bartholomeeusen S, Coenen S (2015) Staphylococcus aureus, including meticillin-resistant Staphylococcus aureus, among general practitioners and their patients: a cross-sectional study. PLoS ONE 10:e0140045
Santos SO, Rocca SM, Hörner R (2016) Colistin resistance in non-fermenting gram-negative bacilli in a university hospital. Braz J Infect Dis 20:649–650
Gniadek TJ, Carroll KC, Simner PJ (2016) Carbapenem-resistant non-glucose-fermenting gram-negative bacilli: the missing piece to the puzzle. J Clin Microbiol 54:1700–1710
Kämpfer P, Avesani V, Janssens M, Charlier J, De Baere T, Vaneechoutte M (2006) Description of Wautersiella falsenii gen. nov., sp. nov., to accommodate clinical isolates phenotypically resembling members of the genera Chryseobacterium and Empedobacter. Int J Syst Evol Microbiol 56:2323–2329
Traglia GM, Dixon C, Chiem K, Almuzara M, Barberis C, Montaña S, Merino C, Mussi MA, Tolmasky ME, Iriarte A, Vay C, Ramírez MS (2015) Draft genome sequence of Empedobacter (Formerly Wautersiella) falsenii comb. nov. Wf282, a strain isolated from a cervical neck abscess. Genome Announc 3:e00235
Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA (2012) SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477
Zerbino DR, Birney E (2008) Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res 18:821–829
Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, Formsma K, Gerdes S, Glass EM, Kubal M, Meyer F, Olsen GJ, Olson R, Osterman AL, Overbeek RA, McNeil LK, Paarmann D, Paczian T, Parrello B, Pusch GD, Reich C, Stevens R, Vassieva O, Vonstein V, Wilke A, Zagnitko O (2008) The RAST Server: rapid annotations using subsystems technology. BMC Genom 9:75
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215:403–410
Gupta SK, Padmanabhan BR, Diene SM, Lopez-Rojas R, Kempf M, Landraud L, Rolain JM (2014) ARG-ANNOT, a new bioinformatic tool to discover antibiotic resistance genes in bacterial genomes. Antimicrob Agents Chemother 58:212–220
Siguier P, Perochon J, Lestrade L, Mahillon J, Chandler M (2006) ISfinder: the reference centre for bacterial insertion sequences. Nucl Acids Res 34:D32–D36
Zhou Y, Liang Y, Lynch K, Dennis JJ, Wishart DS (2011) PHAST: a fast phage search tool. Nucl Acids Res 39:W347–W352
Nielsen H (2017) Predicting secretory proteins with signalp. Methods Mol Biol 1611:59–73
The UniProt Consortium (2017) UniProt: the universal protein knowledgebase. Nucl Acids Res 45:D158–D169
Jones P, Binns D, Chang HY, Fraser M, Li W, McAnulla C et al (2014) InterProScan 5: genome-scale protein function classification. Bioinformatics 30(9):1236–1240
Edgar RC (2004) MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5:113
Li W, Godzik A (2006) Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 22:1658–1659
Talavera G, Castresana J (2007) Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst Biol 56:564–577
Stamatakis A (2014) RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313
Li L, Stoeckert CJ, Roos DS (2003) OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res 13:2178–2189
Contreras-Moreira B, Vinuesa P (2013) GET_HOMOLOGUES, a versatile software package for scalable and robust microbial pangenome analysis. Appl Environ Microbiol 79:7696–7701
Feng Y, Yang P, Wang X, Zong Z (2016) Characterization of Acinetobacter johnsonii isolate XBB1 carrying nine plasmids and encoding NDM-1, OXA-58 and PER-1 by genome sequencing. J Antimicrob Chemother 71:71–75
Sartor AL, Raza MW, Abbasi SA, Day KM, Perry JD, Paterson DL, Sidjabat HE (2014) Molecular epidemiology of NDM-1-producing Enterobacteriaceae and Acinetobacter baumannii isolates from Pakistan. Antimicrob Agents Chemother 58:5589–5593
Bellais S, Girlich D, Karim A, Nordmann P (2002) EBR-1, a novel ambler subclass B1 beta-lactamase from Empedobacter brevis. Antimicrob Agents Chemother 46:3223–3227
Thomas PW, Zheng M, Wu S, Guo H, Liu D, Xu D, Fast W (2011) Characterization of purified New Delhi metallo-β-lactamase-1. Biochemistry 50:10102–10113
Bellais S, Poirel L, Leotard S, Naas T, Nordmann P (2000) Genetic diversity of carbapenem-hydrolyzing metallo-beta-lactamases from Chryseobacterium (Flavobacterium) indologenes. Antimicrob Agents Chemother 44:3028–3034
Cantón R, Morosini MI (2011) Emergence and spread of antibiotic resistance following exposure to antibiotics. FEMS Microbiol Rev 35:977–991
Johnson AP, Woodford N (2013) Global spread of antibiotic resistance: the example of New Delhi metallo-β-lactamase (NDM)-mediated carbapenem resistance. J Med Microbiol 62:499–513
Pfeifer Y, Schlatterer K, Engelmann E, Schiller RA, Frangenberg HR, Stiewe D, Holfelder M, Witte W, Nordmann P, Poirel L (2012) Emergence of OXA-48-type carbapenemase-producing Enterobacteriaceae in German hospitals. Antimicrob Agents Chemother 56:2125–2128
Antonelli A, D’Andrea MM, Vaggelli G, Docquier JD, Rossolini GM (2015) OXA-372, a novel carbapenem-hydrolysing class D β-lactamase from a Citrobacter freundii isolated from a hospital wastewater plant. J Antimicrob Chemother 70:2749–2756
Bellais S, Léotard S, Poirel L, Naas T, Nordmann P (1999) Molecular characterization of a carbapenem-hydrolyzing beta-lactamase from Chryseobacterium (Flavobacterium) indologenes. FEMS Microbiol Lett 171:127–132
Acknowledgements
Secretaría de Ciencia y Técnica de la Universidad de Buenos Aires” (UBACyT) to CV, Buenos Aires, Argentina. GMT, SM, and KC were supported by a Post-doctoral fellowship and doctoral fellowship from CONICET, and grant MHIRT 2T37MD001368 from the National Institute on Minority Health and Health Disparities, National Institute of Health, respectively. AI is a member of the “Sistema Nacional de Investigadores” and “PEDECIBA,” Uruguay.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
284_2018_1498_MOESM1_ESM.pdf
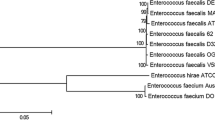
Supplementary material 1 (PDF 77 KB) Maximum likelihood phylogenetic tree of the Flavobacteriaceae family based on 21 conserved ribosomal protein-coding genes. A blue arrow indicates the phylogenetic position of E. falsenii Wf282. Blue branches define the monophyletic group of the closely related genomes of E. falsenii Wf282, bootstrap support is indicated next to nodes. Assembly code of each genome is also indicated. The phylogeny is arbitrary rooted using the midpoint rooting method.
Rights and permissions
About this article
Cite this article
Collins, C., Almuzara, M., Saigo, M. et al. Whole-Genome Analysis of an Extensively Drug-Resistance Empedobacter falsenii Strain Reveals Distinct Features and the Presence of a Novel Metallo-ß-Lactamase (EBR-2). Curr Microbiol 75, 1084–1089 (2018). https://doi.org/10.1007/s00284-018-1498-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00284-018-1498-9