Abstract
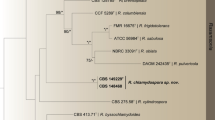
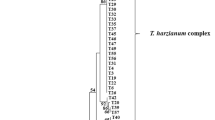
Trichoderma is a dominant component of the soil mycoflora. During the field investigations of northern, central, and southwestern China, three new species in the Stromaticum clade were encountered from soil, and named as T. hebeiense, T. sichuanense, and T. verticillatum. Their phylogenetic positions were determined by analyses of the combined two genes: partial sequences of translation elongation factor 1-alpha and the second largest RNA polymerase subunit-encoding genes. Distinctions between the new species and their close relatives were discussed. Trichoderma hebeiense appeared as a separate terminal branch. The species is distinctive by its oblong conidia and aggregated pustules in culture. Trichoderma sichuanense features in concentric colony and produces numerous clean exudates on aerial mycelium in culture. Trichoderma verticillatum is characterized by its verticillium-like synanamorph and production of abundant chlamydospores. In vitro antagonism towards the new species was tested by dual culture technique.





Similar content being viewed by others
References
Bae SJ, Mohanta TK, Chung JY, Ryu M, Park G, Shim S, Hong SB, Seo H, Bae DW, Bae I (2016) Trichoderma metabolites as biological control agents against Phytophthora pathogens. Biol Control 92:128–138
Bissett J (1984) A revision of the genus Trichoderma. I. Section Longibrachiatum sect. nov. Can J Bot 62:924–931
Bissett J, Szakacs G, Nolan CA, Druzhinina I, Gradinger C, Kubicek CP (2003) New species of Trichoderma from Asia. Can J Bot 81:570–586
Bissett J, Gams W, Jaklitsch WM, Samuels GJ (2015) Accepted Trichoderma names in the year 2015. IMA Fungus 6:263–295
Carbone I, Kohn LM (1999) A method for designing primer sets for speciation studies in filamentous ascomycetes. Mycologia 91:553–556
Chaverri P, Samuels GJ (2013) Evolution of habitat preference and nutrition mode in a cosmopolitan fungal genus with evidence of interkingdom host jumps and major shifts in ecology. Evolution 67:2823–2837
Chen K, Zhuang W-Y (2016) Trichoderma shennongjianum and Trichoderma tibetense, two new soil-inhabiting species in the Strictipile clade. Mycoscience 57:311–319
Cui Y, Wang D, Chai Z-X, Li J-H, Yu B (2014) Biocontrol potential of Trichoderma rossicum:a species new to China. Acta Prataculturae Sinica 23:276–282 (in Chinese)
Cunningham CW (1997) Can three incongruence tests predict when data should be combined? Mol Biol Evol 14:733–740
de Souza JT, Pomella AW, Bowers JH, Pirovani CP, Loguercio LL, Hebbar KP (2006) Genetic and biological diversity of Trichoderma stromaticum, a mycoparasite of the cacao witches’-broom pathogen. Phytopathology 96:61–67
Druzhinina IS, Kopchinskiy AG, Komoń M, Bissett J, Szakacs G, Kubicek CP (2005) An oligonucleotide barcode for species identification in Trichoderma and Hypocrea. Fungal Genet Biol 42:813–828
Harman GE (2000) Myths and dogmas of biocontrol-Changes in perceptions derived from research on Trichoderma harzianum T-22. Plant Dis 84:377–393
Harman GE, Lorito M, Lynch J (2004) Uses of Trichoderma spp. to alleviate or remediate soil and water pollution. Adv Appl Microbiol 56:313
Harman GE (2006) Overview of Mechanisms and Uses of Trichoderma spp. Phytopathology 96:190–194
Hatvani L, Antal Z, Manczinger L, Szekeres A, Druzhinina IS, Kubicek CP, Nagy A, Nagy E, Vagvolgyi C, Kredics L (2007) Green mold diseases of Agaricus and Pleurotus spp. are caused by related but phylogenetically different Trichoderma species. Phytopathology 97:532–537
Jaklitsch WM, Komon M, Kubicek CP, Druzhinina IS (2005) Hypocrea voglmayrii sp. nov. from the Austrian Alps represents a new phylogenetic clade in Hypocrea/Trichoderma. Mycologia 97:1365–1378
Jaklitsch WM (2009) European species of Hypocrea Part I. The green-spored species. Stud Mycol 63:1–91
Jaklitsch WM (2011) European species of Hypocrea part II: species with hyaline ascospores. Fungal Divers 48:1–250
Jaklitsch WM, Voglmayr H (2015) Biodiversity of Trichoderma (Hypocreaceae) in Southern Europe and Macaronesia. Stud Mycol 80:1–87
Kindermann J, El-Ayouti Y, Samuels GJ, Kubicek CP (1998) Phylogeny of the genus Trichoderma based on sequence analysis of the internal transcribed spacer region 1 of the rDNA cluster. Fungal Genet Biol 24:298–309
Klein D, Eveleigh D (1998) Ecology of Trichoderma. In: Kubicek C, Harman G (eds) Trichoderma and Gliocladium. Basic biology, taxonomy and genetics. Taylor and Francis Ltd, London, pp 57–73
Kullnig-Gradinger CM, Szakacs G, Kubicek CP (2002) Phylogeny and evolution of the genus Trichoderma: a multigene approach. Mycol Res 106:757–767
Loguercio LL, de Carvalho AC, Niella GR, De Souza JT, Villela Pomella AW (2009) Selection of Trichoderma stromaticum isolates for efficient biological control of witches’ broom disease in cacao. Biol Control 51:130–139
Montoya QV, Meirelles LA, Chaverri P, Rodrigues A (2016) Unraveling Trichoderma species in the attine ant environment: description of three new taxa. Anton Leeuw Int J G 109:633–651
Nirenberg H (1976) Untersuchungen über die morphologische und biologische Differenzierung in der Fusarium-Sektion Liseola. Mitt aus der Biol Bundesanst für Land- u Forstwirtsch, Berlin-Dahlem 169:1–117
Nylander JAA (2004) MrModeltest v2. Program distributed by the author. Evolutionary Biology Centre, Uppsala University, Uppsala
Page R (1996) TreeView: an application to display phylogenetic trees on personal computers. Comput Appl Biosci 12:357–358
Park MS, Bae KS, Yu SH (2006) Two new species of Trichoderma associated with green mold of oyster mushroom cultivation in Korea. Mycobiology 34:111–113
Persoon CH (1794) Dispositio methodica fungorum. Neues Magazin für die Botanik 1: 81–128
Polanco R, Pino C, Besoain X, Montealegre J, Pérez LM (2015) Enhanced secretion of biocontrol enzymes by Trichoderma harzianum mutant strains in the presence of Rhizoctonia solani cell walls. Ciencia e Investigación Agraria 42:243–250
Qin W-T, Zhuang W-Y (2016) Two new hyaline-ascospored species of Trichoderma and their phylogenetic positions. Mycologia 108:205–214
Qin W-T, Zhuang W-Y (2016) Seven wood-inhabiting new species of the genus Trichoderma (Fungi, Ascomycota) in Viride clade. Sci Rep-UK 6:27074
Qin W-T, Zhuang W-Y (2016) Four new species of Trichoderma with hyaline ascospores from central China. Mycol Prog 15:811–825
Qin W-T, Chen K, Zhuang W-Y (2016) Five Trichoderma species new to China and notes on two other widespread species. Mycosystema 35:994–1007 (in Chinese)
Rifai MA (1969) Revision of the genus Trichoderma. Mycological Papers 116:1–56
Ronquist F, Huelsenbeck JP (2003) MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19:1572–1574
Samuels GJ, Pardo-Schultheiss R, Hebbar KP, Lumsden RD, Bastos CN, Costa JC, Bezerra JL (2000) Trichoderma stromaticum sp. nov., a parasite of the cacao witches broom pathogen. Mycol Res 104:760–764
Samuels GJ, Dodd SL, Gams W, Castlebury LA, Petrini O (2002) Trichoderma species associated with the green mold epidemic of commercially grown Agaricus bisporus. Mycologia 94:146–170
Samuels GJ, Ismaiel A, de Souza J, Chaverri P (2012) Trichoderma stromaticum and its overseas relatives. Mycol Prog 11:215–254
Sandoval-Denis M, Sutton DA, Cano-Lira JF, Gené J, Fothergill AW, Wiederhold NP, Guarro J (2014) Phylogeny of the clinically relevant species of the emerging fungus Trichoderma and their antifungal susceptibilities. J Clin Microbiol 52:2112–2125
Schuster A, Schmoll M (2010) Biology and biotechnology of Trichoderma. Appl Microbiol Biot 87:787–799
Stamatakis A (2006) RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22:2688–2690
Sun R-Y, Liu Z-C, Fu K, Fan L, Chen J (2012) Trichoderma biodiversity in China. J Appl Genet 53:343–354
Swofford DL (2002) PAUP* 4.0b10: phylogenetic analysis using parsimony (* and other methods). Sunderland, Massachusetts: Sinauer Associates
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucl Acids Res 25:4876–4882
Wang L, Zhuang W-Y (2004) Designing primer sets for amplification of partial calmodulin genes from penicillia. Mycosystema 23:466–473
White TJ, Bruns T, Lee S, Taylor J (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfland DH, Sninsky JJ, White TJ (eds) PCR protocols: a guide to methods and applications. Academic Press, San Diego, pp 315–322
Zhu Z-X, Zhuang W-Y (2014) Two new species of Trichoderma (Hypocreaceae) from China. Mycosystema 33:1168–1174
Acknowledgements
We thank Dr. Long Wang for providing the soil samples from Sichuan, Dr. Gary Samuels for linguistic corrections and valuable suggestions, and Ms. Xia Song for technique assistance. This project was supported by the National Natural Science Foundation of China (No. 31570018) and Ministry of Science and Technology of China for Fundamental Research (No. 2013FY110400).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Chen, K., Zhuang, WY. Three New Soil-inhabiting Species of Trichoderma in the Stromaticum Clade with Test of Their Antagonism to Pathogens. Curr Microbiol 74, 1049–1060 (2017). https://doi.org/10.1007/s00284-017-1282-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00284-017-1282-2