Abstract
Purpose
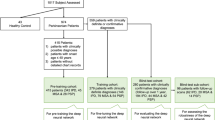
This study investigated the potential of deep convolutional neural networks (CNN) for automatic classification of FP-CIT SPECT in multi-site or multi-camera settings with variable image characteristics.
Methods
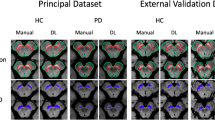
The study included FP-CIT SPECT of 645 subjects from the Parkinson’s Progression Marker Initiative (PPMI), 207 healthy controls, and 438 Parkinson’s disease patients. SPECT images were smoothed with an isotropic 18-mm Gaussian kernel resulting in 3 different PPMI settings: (i) original (unsmoothed), (ii) smoothed, and (iii) mixed setting comprising all original and all smoothed images. A deep CNN with 2,872,642 parameters was trained, validated, and tested separately for each setting using 10 random splits with 60/20/20% allocation to training/validation/test sample. The putaminal specific binding ratio (SBR) was computed using a standard anatomical ROI predefined in MNI space (AAL atlas) or using the hottest voxels (HV) analysis. Both SBR measures were trained (ROC analysis, Youden criterion) using the same random splits as for the CNN. CNN and SBR trained in the mixed PPMI setting were also tested in an independent sample from clinical routine patient care (149 with non-neurodegenerative and 149 with neurodegenerative parkinsonian syndrome).
Results
Both SBR measures performed worse in the mixed PPMI setting compared to the pure PPMI settings (e.g., AAL-SBR accuracy = 0.900 ± 0.029 in the mixed setting versus 0.957 ± 0.017 and 0.952 ± 0.015 in original and smoothed setting, both p < 0.01). In contrast, the CNN showed similar accuracy in all PPMI settings (0.967 ± 0.018, 0.972 ± 0.014, and 0.955 ± 0.009 in mixed, original, and smoothed setting). Similar results were obtained in the clinical sample. After training in the mixed PPMI setting, only the CNN provided acceptable performance in the clinical sample.
Conclusions
These findings provide proof of concept that a deep CNN can be trained to be robust with respect to variable site-, camera-, or scan-specific image characteristics without a large loss of diagnostic accuracy compared with mono-site/mono-camera settings. We hypothesize that a single CNN can be used to support the interpretation of FP-CIT SPECT at many different sites using different acquisition hardware and/or reconstruction software with only minor harmonization of acquisition and reconstruction protocols.




Similar content being viewed by others
References
Booij J, Speelman JD, Horstink MW, Wolters EC. The clinical benefit of imaging striatal dopamine transporters with [123I]FP-CIT SPET in differentiating patients with presynaptic parkinsonism from those with other forms of parkinsonism. Eur J Nucl Med. 2001;28:266–72.
Darcourt J, Booij J, Tatsch K, Varrone A, Vander Borght T, Kapucu OL, et al. EANM procedure guidelines for brain neurotransmission SPECT using (123)I-labelled dopamine transporter ligands, version 2. Eur J Nucl Med Mol Imaging. 2010;37:443–50. https://doi.org/10.1007/s00259-009-1267-x.
Tatsch K, Poepperl G. Nigrostriatal dopamine terminal imaging with dopamine transporter SPECT: an update. J Nucl Med. 2013;54:1331–8. https://doi.org/10.2967/jnumed.112.105379.
Van Laere K, Everaert L, Annemans L, Gonce M, Vandenberghe W, Vander Borght T. The cost effectiveness of 123I-FP-CIT SPECT imaging in patients with an uncertain clinical diagnosis of parkinsonism. Eur J Nucl Med Mol Imaging. 2008;35:1367–76. https://doi.org/10.1007/s00259-008-0777-2.
O'Brien JT, Oertel WH, McKeith IG, Grosset DG, Walker Z, Tatsch K, et al. Is ioflupane I123 injection diagnostically effective in patients with movement disorders and dementia? Pooled analysis of four clinical trials. BMJ Open. 2014;4:ARTN e005122. https://doi.org/10.1136/bmjopen-2014-005122.
Seibyl JP, Kupsch A, Booij J, Grosset DG, Costa DC, Hauser RA, et al. Individual-reader diagnostic performance and between-reader agreement in assessment of subjects with Parkinsonian syndrome or dementia using 123I-ioflupane injection (DaTscan) imaging. J Nucl Med. 2014;55:1288–96. https://doi.org/10.2967/jnumed.114.140228.
Booij J, Dubroff J, Pryma D, Yu J, Agarwal R, Lakhani P, et al. Diagnostic performance of the visual reading of I-123-ioflupane SPECT images with or without quantification in patients with movement disorders or dementia. J Nucl Med. 2017;58:1821–6. https://doi.org/10.2967/jnumed.116.189266.
Soderlund TA, Dickson JC, Prvulovich E, Ben-Haim S, Kemp P, Booij J, et al. Value of semiquantitative analysis for clinical reporting of 123I-2-beta-carbomethoxy-3beta-(4-iodophenyl)-N-(3-fluoropropyl)nortropane SPECT studies. J Nucl Med. 2013;54:714–22. https://doi.org/10.2967/jnumed.112.110106.
Badiavas K, Molyvda E, Iakovou I, Tsolaki M, Psarrakos K, Karatzas N. SPECT imaging evaluation in movement disorders: far beyond visual assessment. Eur J Nucl Med Mol Imaging. 2011;38:764–73. https://doi.org/10.1007/s00259-010-1664-1.
Tatsch K, Poepperl G. Quantitative approaches to dopaminergic brain imaging. Q J Nucl Med Mol Imaging. 2012;56:27–38.
Oliveira FPM, Faria DB, Costa DC, Castelo-Branco M, Tavares J. Extraction, selection and comparison of features for an effective automated computer-aided diagnosis of Parkinson's disease based on [(123)I]FP-CIT SPECT images. Eur J Nucl Med Mol Imaging. 2018;45:1052–62. https://doi.org/10.1007/s00259-017-3918-7.
Nobili F, Naseri M, De Carli F, Asenbaum S, Booij J, Darcourt J, et al. Automatic semi-quantification of [123I]FP-CIT SPECT scans in healthy volunteers using BasGan version 2: results from the ENC-DAT database. Eur J Nucl Med Mol Imaging. 2013;40:565–73. https://doi.org/10.1007/s00259-012-2304-8.
Albert NL, Unterrainer M, Diemling M, Xiong GM, Bartenstein P, Koch W, et al. Implementation of the European multicentre database of healthy controls for [I-123]FP-CIT SPECT increases diagnostic accuracy in patients with clinically uncertain parkinsonian syndromes. Eur J Nucl Med Mol Imaging. 2016;43:1315–22. https://doi.org/10.1007/s00259-015-3304-2.
Dickson JC, Tossici-Bolt L, Sera T, Booij J, Ziebell M, Morbelli S, et al. The impact of reconstruction and scanner characterisation on the diagnostic capability of a normal database for [123I]FP-CIT SPECT imaging. EJNMMI Res. 2017;7:10. https://doi.org/10.1186/s13550-016-0253-0.
Fujita M, Varrone A, Kim KM, Watabe H, Zoghbi SS, Baldwin RM, et al. Effect of scatter correction in the measurement of striatal and extrastriatal dopamine D2 receptors using (123)Iepidepride SPECT. J Nucl Med. 2001;42:217.
Lange C, Seese A, Schwarzenbock S, Steinhoff K, Umland-Seidler B, Krause BJ, et al. CT-based attenuation correction in I-123-ioflupane SPECT. PLoS One. 2014;9:e108328. https://doi.org/10.1371/journal.pone.0108328.
Meyer PT, Sattler B, Lincke T, Seese A, Sabri O. Investigating dopaminergic neurotransmission with I-123-FP-CIT SPECT: comparability of modern SPECT systems. J Nucl Med. 2003;44:839–45.
Tossici-Bolt L, Dickson JC, Sera T, Booij J, Asenbaun-Nan S, Bagnara MC, et al. [123I]FP-CIT ENC-DAT normal database: the impact of the reconstruction and quantification methods. EJNMMI Phys. 2017;4:8. https://doi.org/10.1186/s40658-017-0175-6.
Tossici-Bolt L, Dickson JC, Sera T, de Nijs R, Bagnara MC, Jonsson C, et al. Calibration of gamma camera systems for a multicentre European (1)(2)(3)I-FP-CIT SPECT normal database. Eur J Nucl Med Mol Imaging. 2011;38:1529–40. https://doi.org/10.1007/s00259-011-1801-5.
Varrone A, Dickson JC, Tossici-Bolt L, Sera T, Asenbaum S, Booij J, et al. European multicentre database of healthy controls for [123I]FP-CIT SPECT (ENC-DAT): age-related effects, gender differences and evaluation of different methods of analysis. Eur J Nucl Med Mol Imaging. 2013;40:213–27. https://doi.org/10.1007/s00259-012-2276-8.
Buchert R, Kluge A, Tossici-Bolt L, Dickson J, Bronzel M, Lange C, et al. Reduction in camera-specific variability in [(123)I]FP-CIT SPECT outcome measures by image reconstruction optimized for multisite settings: impact on age-dependence of the specific binding ratio in the ENC-DAT database of healthy controls. Eur J Nucl Med Mol Imaging. 2016;43:1323–36. https://doi.org/10.1007/s00259-016-3309-5.
Koch W, Bartenstein P, la Fougere C. Radius dependence of FP-CIT quantification: a Monte Carlo-based simulation study. Ann Nucl Med. 2014;28:103–11. https://doi.org/10.1007/s12149-013-0789-2.
Koch W, Mustafa M, Zach C, Tatsch K. Influence of movement on FP-CIT SPECT quantification: a Monte Carlo based simulation. Nucl Med Commun. 2007;28:603–14. https://doi.org/10.1097/MNM.0b013e328273bc6f.
LeCun Y, Bengio Y, Hinton G. Deep learning. Nature. 2015;521:436–44. https://doi.org/10.1038/nature14539.
Litjens G, Kooi T, Bejnordi BE, Setio AAA, Ciompi F, Ghafoorian M, et al. A survey on deep learning in medical image analysis. Med Image Anal. 2017;42:60–88. https://doi.org/10.1016/j.media.2017.07.005.
Choi H, Jin KH. Predicting cognitive decline with deep learning of brain metabolism and amyloid imaging. Behav Brain Res. 2018;344:103–9. https://doi.org/10.1016/j.bbr.2018.02.017.
Ding Y, Sohn JH, Kawczynski MG, Trivedi H, Harnish R, Jenkins NW, et al. A deep learning model to predict a diagnosis of Alzheimer disease by using (18)F-FDG PET of the brain. Radiology. 2018:180958. https://doi.org/10.1148/radiol.2018180958.
Liu M, Cheng D, Yan W. Alzheimer’s disease neuroimaging I. classification of Alzheimer’s disease by combination of convolutional and recurrent neural networks using FDG-PET images. Front Neuroinform. 2018;12:35. https://doi.org/10.3389/fninf.2018.00035.
Lu D, Popuri K, Ding GW, Balachandar R, Beg MF. Multiscale deep neural network based analysis of FDG-PET images for the early diagnosis of Alzheimer’s disease. Med Image Anal. 2018;46:26–34. https://doi.org/10.1016/j.media.2018.02.002.
Segovia F, Gorriz JM, Ramirez J, Martinez-Murcia FJ, Garcia-Perez M. Using deep neural networks along with dimensionality reduction techniques to assist the diagnosis of neurodegenerative disorders. Log J IGPL. 2018;26:618–28. https://doi.org/10.1093/jigpal/jzy026.
Shi J, Zheng X, Li Y, Zhang Q, Ying S. Multimodal neuroimaging feature learning with multimodal stacked deep polynomial networks for diagnosis of Alzheimer’s disease. IEEE J Biomed Health Inform. 2018;22:173–83. https://doi.org/10.1109/jbhi.2017.2655720.
Zhou T, Thung KH, Zhu X, Shen D. Effective feature learning and fusion of multimodality data using stage-wise deep neural network for dementia diagnosis. Hum Brain Mapp. 2018. https://doi.org/10.1002/hbm.24428.
Hirata K, Takeuchi W, Yamaguchi S, Kobayashi H, Terasaka S, Toyonaga T, et al. Convolutional neural network can help differentiate FDG PET images of brain tumor between glioblastoma and primary central nervous system lymphoma. J Nucl Med. 2016;57.
Blanc-Durand P, Van der Gucht A, Schaefer N, Itti E, Prior JO. Automatic lesion detection and segmentation of F-18-FET PET in gliomas: a full 3D U-net convolutional neural network study. PLoS One. 2018;13:ARTN e0195798. https://doi.org/10.1371/journal.pone.0195798.
Kang SK, Seo S, Shin SA, Byun MS, Lee DY, Kim YK, et al. Adaptive template generation for amyloid PET using a deep learning approach. Hum Brain Mapp. 2018;39:3769–78. https://doi.org/10.1002/hbm.24210.
Xiang L, Qiao Y, Nie D, An L, Lin WL, Wang Q, et al. Deep auto-context convolutional neural networks for standard-dose PET image estimation from low-dose PET/MRI. Neurocomputing. 2017;267:406–16. https://doi.org/10.1016/j.neucom.2017.06.048.
Wang Y, Yu BT, Wang L, Zu C, Lalush DS, Lin WL, et al. 3D conditional generative adversarial networks for high-quality PET image estimation at low dose. Neuroimage. 2018;174:550–62. https://doi.org/10.1016/j.neuroimage.2018.03.045.
Li RJ, Zhang WL, Suk HI, Wang L, Li J, Shen DG, et al. Deep learning based imaging data completion for improved brain disease diagnosis. Lect Notes Comput Sc. 2014;8675:305–12.
Liu F, Jang H, Kijowski R, Bradshaw T, McMillan AB. Deep learning MR imaging-based attenuation correction for PET/MR imaging. Radiology. 2018;286:676–84. https://doi.org/10.1148/radiol.2017170700.
Parkinson Progression Marker I. The Parkinson progression marker initiative (PPMI). Prog Neurobiol. 2011;95:629–35. https://doi.org/10.1016/j.pneurobio.2011.09.005.
Djang DS, Janssen MJ, Bohnen N, Booij J, Henderson TA, Herholz K, et al. SNM practice guideline for dopamine transporter imaging with 123I-ioflupane SPECT 1.0. J Nucl Med. 2012;53:154–63. https://doi.org/10.2967/jnumed.111.100784.
Yokoyama K, Imabayashi E, Sumida K, Sone D, Kimura Y, Sato N, et al. Computed-tomography-guided anatomic standardization for quantitative assessment of dopamine transporter SPECT. Eur J Nucl Med Mol Imaging. 2017;44:366–72. https://doi.org/10.1007/s00259-016-3496-0.
Kupitz D, Apostolova I, Lange C, Ulrich G, Amthauer H, Brenner W, et al. Global scaling for semi-quantitative analysis in FP-CIT SPECT. Nuklearmed-Nucl Med. 2014;53:234–41. https://doi.org/10.3413/Nukmed-0659-14-04.
Koch W, Unterrainer M, Xiong G, Bartenstein P, Diemling M, Varrone A, et al. Extrastriatal binding of [(1)(2)(3)I]FP-CIT in the thalamus and pons: gender and age dependencies assessed in a European multicentre database of healthy controls. Eur J Nucl Med Mol Imaging. 2014;41:1938–46. https://doi.org/10.1007/s00259-014-2785-8.
Tzourio-Mazoyer N, Landeau B, Papathanassiou D, Crivello F, Etard O, Delcroix N, et al. Automated anatomical labeling of activations in SPM using a macroscopic anatomical parcellation of the MNI MRI single-subject brain. Neuroimage. 2002;15:273–89. https://doi.org/10.1006/nimg.2001.0978.
Youden WJ. Index for rating diagnostic tests. Cancer. 1950;3:32–5.
Patel AB, Nguyen T, Baraniuk RG. A probabilistic framework for deep learning. Adv Neur In. 2016;29.
Szegedy C, Vanhouke V, Ioffe S, Shlens J, Wojna Z. Rethinking the inception architecture for computer vision. In: IEEE conference on computer vision and pattern recognition; 2015. p. 2818–26.
Russakovsky O, Deng J, Su H, Krause J, Sateesh S, Ma S, et al. ImageNet large scale visual recognition challenge. J Comput Vision. 2014;115:1–42.
Choi H, Ha S, Im HJ, Paek SH, Lee DS. Refining diagnosis of Parkinson’s disease with deep learning-based interpretation of dopamine transporter imaging. Neuroimage Clin. 2017;16:586–94. https://doi.org/10.1016/j.nicl.2017.09.010.
Kim DH, Wit H, Thurston M. Artificial intelligence in the diagnosis of Parkinson’s disease from ioflupane-123 single-photon emission computed tomography dopamine transporter scans using transfer learning. Nucl Med Commun. 2018;39:887–93. https://doi.org/10.1097/MNM.0000000000000890.
Martinez-Murcia FJ, Gorriz JM, Ramirez J, Ortiz A. Convolutional neural networks for neuroimaging in Parkinson’s disease: is preprocessing needed? Int J Neural Syst. 2018:1850035. https://doi.org/10.1142/S0129065718500351.
Giorgio A, De Stefano N. Clinical use of brain volumetry. J Magn Reson Imaging. 2013;37:1. https://doi.org/10.1002/jmri.23671.
Han X, Jovicich J, Salat D, van der Kouwe A, Quinn B, Czanner S, et al. Reliability of MRI-derived measurements of human cerebral cortical thickness: the effects of field strength, scanner upgrade and manufacturer. Neuroimage. 2006;32:180–94. https://doi.org/10.1016/j.neuroimage.2006.02.051.
Kouw WM, Loog M, Bartels LW, Mendrik AM. MR acquisition invariant representation learning. arXiv. 2018;arXiv:1709.07944v2.
Ghafoorian M, Mehrtash A, Kapur T, Karssemeijer N, Marchiori E, Pesteie M, et al. Transfer learning for domain adaptation in MRI: application in brain lesion segmentation. arXiv. 2017; https://arxiv.org/pdf/1702.07841.pdf.
Hosseini-Asl E, Keynton R, El-Baz A. Alzheimer’s disease diagnostics by adaption of 3D convolutional network. arXiv. 2016; https://arxiv.org/pdf/1702.07841.pdf.
Valverde S, Salem M, Cabezas M, Pareto D, Vilanova JC, Ramio-Torrenta L, et al. One-shot domain adaptation in multiple sclerosis lesion segmentation using convolutional neural networks. Neuroimage Clin. 2019;21:101638. https://doi.org/10.1016/j.nicl.2018.101638.
Joshi A, Koeppe RA, Fessler JA. Reducing between scanner differences in multi-center PET studies. Neuroimage. 2009;46:154–9. https://doi.org/10.1016/j.neuroimage.2009.01.057.
Acknowledgments
PPMI—a public-private partnership—is funded by the Michael J. Fox Foundation for Parkinson’s Research and funding partners including the following: Abbvie, Avid Radiopharmaceuticals, Biogen, BioLegend, Bristol-Myers Squibb, GE Healthcare, Genentech, GlaxoSmithKline, Lilly, Lundbeck, Merck, Meso Scale Discovery, Pfizer, Piramal, Roche, Sanofi Genzyme, Servier, Takeda, Teva, and UCB. For up-to-date information about all of the PPMI funding partners, visit www.ppmi-info.org/fundingpartners.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Ethical approval
Waiver of informed consent for the retrospective analyses of the anonymized clinical data in this study was obtained from the ethics review board of the general medical council of the state of Hamburg, Germany. All procedures performed in this study were in accordance with the ethical standards of the ethics review board of the general medical council of the state of Hamburg, Germany, and with the 1964 Helsinki declaration and its later amendments.
Conflict of interest
F.M. is employee of Nvidia, J. K is employee of Jung Diagnostics, and M.E. is employee of Pinax Pharma. This did not influence the content of this manuscript, neither directly nor indirectly. There is no actual or potential conflict of interest for any of the other authors.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This article is part of the Topical Collection on Advanced Image Analyses (Radiomics and Artificial Intelligence)
Electronic supplementary material
ESM 1
(DOCX 26 kb)
Rights and permissions
About this article
Cite this article
Wenzel, M., Milletari, F., Krüger, J. et al. Automatic classification of dopamine transporter SPECT: deep convolutional neural networks can be trained to be robust with respect to variable image characteristics. Eur J Nucl Med Mol Imaging 46, 2800–2811 (2019). https://doi.org/10.1007/s00259-019-04502-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00259-019-04502-5