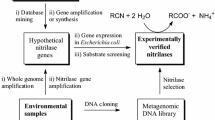
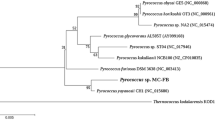
Abstract
Commercially, nitrilases are valuable biocatalysts capable of converting a diverse range of nitriles to carboxylic acids for the greener synthesis of chemicals and pharmaceuticals. Nitrilases are widespread in nature and are both important components of metabolic pathways and a response to environmental factors such as natural or manmade nitriles. Nitrilases are often grouped together on a genome in specific gene clusters that reflect these metabolic functions. Although nitrilase induction systems are still poorly understood, it is known that a powerful Rhodococcal transcription regulator system permits accumulation of intracellular nitrilase of up to 30–40% of total soluble protein in wild type Rhodococcous rhodochrous and host Streptomyces strains. Nitrilase expression inducer molecules encompass a broad range of aliphatic, aromatic and heteroaromatic nitriles, as well as some secondary and tertiary amides that are resistant to nitrilase degradation.






Similar content being viewed by others
Notes
Oddly, the nitA gene product is labelled as an aliphatic nitrilase in UniProt (UniProtKB Q03217: NRL2_RHORH)
References
Almatawah QA, Cowan DA (1999) Thermostable nitrilase catalyzed production of nicotinic acid from 3-cyanopyridine. Enzym Microb Technol 25:718–724
Almatawah QA, Cramp R, Cowan DA (1999) Characterization of an inducible nitrilase from a thermophilic bacillus. Extremophiles 3:283–291
Alonso FOM, Oestreicher EG, Antunes OAC (2008) Production of enantiomerically pure D-phenylglycine using Pseudomonas aeruginosa 10145 as biocatalyst. Braz J Chem Eng 25:1–8
Bandyopadhyay AK, Nagasawa T, Asano Y, Fujishiro K, Tani Y, Yamada H (1986) Purification and characterisation of benzonitrilases from Arthrobacter sp. strain J-1. Appl Environ Microbiol 51:302–306
Banerjee A, Kaul P, Banerjee UC (2006) Purification and characterization of an enantioselective arylacetonitrilase from Pseudomonas putida. Arch Microbiol 184:407–418
Banerjee A, Dubey S, Kaul P, Barse B, Piotrowski M, Banerjee UC (2009) Enantioselective nitrilase from Pseudomonas putida: cloning, heterologous expression, and bioreactor studies. Mol Biotechnol 41:35–41
Bayer S, Birkemeyer C, Ballschmite M (2011) A nitrilase from a metagenomic library acts regioselectively on aliphatic dinitriles. Appl Microbiol Biotechnol 89:91–98
Beck A, Divakar PK, Zhang N, Molina MC, Struwe L (2015) Evidence of ancient horizontal gene transfer between fungi and the terrestrial alga Trebouxia. Org Divers Evol 15:235–248
Bekker V, Dodd A, Brady D, Rumbold K (2014) Tools for metabolic engineering in Streptomyces. Bioengineered 5:293–299
Bhalla TC, Miura A, Wakamoto A, Ohba Y, Furuhasi K (1992) Asymmetric hydrolysis of α-amino nitriles by a nitrilase of Rhodococcus rhodochrous PA-34. Appl Microbiol Biotechnol 37:184–190
Bork P, Koonin EV (1994) A new family of carbon-nitrogen hydrolases. Protein Sci 3:1344–1346
Brenner C (2002) Catalysis in the nitrilase superfamily. Curr Opin Struct Biol 12:775–782
Brunner S, Eppinger E, Fischer S, Gröning J, Stolz A (2018) Conversion of aliphatic nitriles by the arylacetonitrilase from Pseudomonas fluorescens EBC191. World J Microbiol Biotechnol 34:91
Chen K, Huang L, Xu C, Liu X, He J, Zinder SH, Li S, Jiang J (2013) Molecular characterization of the enzymes involved in the degradation of a brominated aromatic herbicide. Mol Microbiol 89:1121–1139
Chhiba-Govindjee VP, Mathiba K, van der Westhuyzen CW, Steenkamp P, Rashamuse JK, Stoychev S, Bode ML, Brady D (2018) Dimethylformamide is a novel nitrilase inducer in Rhodococcus rhodochrous. Appl Microbiol Biotechnol 102:10055–10065
Copely SD (2017) Shining a light on enzyme promiscuity. Curr Opin Struct Biol 47:167–175
Dennet GV, Blamey JM (2016) A new thermophilic nitrilase from an Antarctic hyperthermophilic microorganism. Front Bioeng Biotechnol 4:5
Dhillon JK, Shivaraman N (1999) Biodegradation of cyanide compounds by a Pseudomonas species (S1). Can J Microbiol 45:201–208
Dooley-Cullinane T-M, O’Reilly C, Weiner DP AB, O'Neill D, Owens E, O'Meara D, Coffey L (2018) The use of clade-specific PCR assays to identify novel nitrilase genes from environmental isolates. Microbiol Open 2018:e700
Estepa J, Luque-Almagro VM, Manso I, Paz Escribano M, Martínez-Luque M, Castillo F, Moreno-Vivián C, Dolores Roldán M (2012) The nit1C gene cluster of Pseudomonas pseudoalcaligenes CECT5344 involved in assimilation of nitriles is essential for growth on cyanide. Environ Microbiol Rep 4:326–334
Expasy Enzyme database (2019) https://enzyme.expasy.org/cgi-bin/enzyme/enzyme-search-de. Accessed 01 Feb 2019
Fan H, Chen L, Sun H, Wang H, Liu Q, Ren Y, Wei D (2017) Characterization of a novel nitrilase, BGC4, from Paraburkholderia graminis showing wide-spectrum substrate specificity, a potential versatile biocatalyst for the degradation of nitriles. Biotechnol Lett 39:1725–1731
Ferrer M, Martínez-Martínez M, Bargiela R, Streit WR, Golyshina OV, Golyshin PN (2016) Estimating the success of enzyme bioprospecting through metagenomics: current status and future trends. Microb Biotechnol 9:22–34
Fischer-Colbrie G, Matama T, Heumann S, Martinkova L, Paulo AC, Guebitz G (2007) Surface hydrolysis of polyacrylonitrile with nitrile hydrolysing enzymes from Micrococcus luteus BST20. J Biotechnol 129:62–68
Gallegos MT, Schleif R, Bairoch A, Hofmann K, Ramos JL (1997) AraC/XylS family of transcriptional regulators. Microbiol Molec Biol Rev 61:393–410
Gavagan JE, DiCosimo R, Eisenberg A, Folsom PW FSK, Hahn EC, Shneide KJ, Fallon RD (1999) A Gram-negative bacterium producing a heat-stable nitrilase highly active on aliphatic dinitriles. Appl Microbiol Biotechnol 52:654–659
Goldlust A, Bohak Z (1989) Induction, purification, and characterization of the nitrilase of Fusarium oxysporum f. sp. melonis. Biotechnol Appl Biochem 11:581–601
Gong J-S, Li H, Zhu X-Y, Lu Z-M, Wu Y, Shi J-S, Xu Z-H (2012a) Fungal His-tagged nitrilase from Gibberella intermedia: gene cloning, heterologous expression and biochemical properties. PLoS One 7(11):e50622
Gong J-S, Lu Z-M, Li H, Shi J-S, Zhou Z-M, Xu Z-H (2012b) Nitrilases in nitrile biocatalysis: recent progress and forthcoming research. Microb Cell Factories 11:1–18
Günther J, Irmisch S, Lackus ND, Reichelt M, Köllner GJ (2018) The nitrilase PtNIT1 catabolizes herbivore-induced nitriles in Populus triochocarpa. BMC Plant Biol 18:251
Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser 41:95–98
Hann EC, Sigmund AE, Fager SK, Cooling FB, Gavagan JE, Bramucci MG, Chauhan S, Payne MS, DiCosimo R (2004) Regioselective biocatalytic hydrolysis of (E,Z)-2-methyl-2-butenenitrile for production of (E)-2-methyl-2-butenoic acid. Tetrahedron 60:577–581
Harper DB (1977a) Microbial metabolism of aromatic nitriles: enzymology of C-N cleavage by Nocardia sp. (Rhodococcus group) NCIB 11216. Biochem J 165:309–319
Harper DB (1977b) Fungal degradation of aromatic nitriles enzymology of C-N cleavage by Fusarium solani. J Biochem 167:685–692
Harper DB (1985) Characterization of a nitrilase from Nocardia sp. (Rhodochrous group) N.C.I.B. 11215, using p-hydroxybenzonitrile as sole carbon source. Int J BioChemiPhysics 17:677–683
Hashimoto Y (2007) Development of novel expression systems for actinomycetes. Actinomycetologica 21:70–75
Heinemann U, Engels D, Burger S, Kiziak C, Mattes R, Stolz A (2003) Cloning of a nitrilase gene from the cyanobacterium Synechocystis sp. strain PCC6803 and heterologous expression and characterization of the encoded protein. Appl Environ Microbiol 69:4359–4366
Herai S, Hashimoto Y, Higashibata H, Maseda H, Ikeda H, Ōmura S, Kobayashi M (2004) Hyper-inducible expression system for Streptomycetes. Proc Nat Acad Sci 101:14031–14035
Hook RH, Robinson WG (1964) Ricinine nitrilase: II. Purification and properties. J Biol Chem 239:4263–4267
Howden AJM, Preston GM (2009) Nitrilase enzymes and their role in plant-microbe interactions. Microbial Biotechnol 2:441–451
Howden AJM, Rico A, Mentlak T, Miguet L, Preston GM (2009a) Pseudomonas syringae pv. syringae B728a hydrolyses indole-3-acetonitrile to the plant hormone indole-3-acetic acid. Mol Plant Pathol 10:857–865
Howden AJM, Harrison CJ, Preston GM (2009b) A conserved mechanism for nitrile metabolism in bacteria and plants. Plant J 57:243–253
Hoyle AJ, Bunch AW, Knowles CJ (1998) The nitrilases of Rhodococcus rhodochrous NCIMB 11216. Enzym Microb Technol 23:475–482
Jones LB, Ghosh P, Lee J-H, Chou C-N, Kunz DA (2018) Linkage of the Nit1C gene cluster to bacterial cyanide assimilation as a nitrogen source. Microbiology 164:956–968
Jyoti BK, Chauhan K, Attri C, Seth A (2017) Improving stability and reusability of Rhodococcus pyridinivorans NIT-36 nitrilase by whole cell immobilization using chitosan. IntJ Biol Macromol 103:8–15
Kamal A, Kumar MS, Kumar CG, Shaik TB (2011) Bioconversion of acrylonitrile to acrylic acid by Rhodococcus ruber strain AKSH-84. J Microbiol Biotechnol 21:37–42
Kaplan O, Nikolau K, Pisvejcova CA, Martinkova L (2006a) Hydrolysis of nitriles and amides by filamentous fungi. Enzym Microb Technol 38:260–264
Kaplan O, Vejvoda V, Charvátová-Pišvejcová A, Martínková L (2006b) Hyperinduction of nitrilases in filamentous fungi. J Ind Microbiol Biotechnol 33:891–896
Kato Y, Nakamura K, Sakiyama H, Mayhew SG, Asano Y (2000) Novel heme-containing lyase, phenylacetaldoxime dehydratase from Bacillus sp, strain OxB-1: purification, characterization, and molecular cloning of the gene. Biochem 39:800–809
Kaur G, Soni P, Tewari R, Sharma R (2014) Isolation and characterization of a nitrile-hydrolysing bacterium Isoptericola variabilis RGT01. Indian J Microbiol 54:232–238
Khandelwal AK, Nigam VK, Chaudary B, Mohan MK, Ghosh P (2007) Optimization of nitrilase production from a new thermophilic isolate. J Chem Technol Biotechnol 82:646–651
Kiziak C, Conradt D, Stolz A, Mattes R, Klein J (2005) Nitrilase from Pseudomonas fluorescens EBC191: cloning and heterologous expression of the gene and bio-chemical characterization of the recombinant enzyme. Microbiology 151:3639–3648
Kobayashi M, Shimizu S (1994) Versatile nitrilases: nitrile-hydrolysing enzymes. FEMS Microbiol Lett 120:217–224
Kobayashi M, Nagasawa T, Yamada H (1989) Nitrilase of Rhodococcus rhodochrous J1. Purification and characterization. Eur J Biochem 182:349–356
Kobayashi M, Yanaka N, Nagasawa T, Yamada H (1990) Purification and characterization of a novel nitrilase of Rhodococcus rhodochrous K22 that acts on aliphatic nitriles. J Bacteriol 17:4807–4815
Kobayashi M, Yanaka N, Nagasawa T, Yamada H (1991) Hyperinduction of an aliphatic nitrilase by Rhodococcus rhodochrous K22. FEMS Microbiol Lett 77:121–123
Kobayashi M, Komeda H, Yanaka N, Nagasawa T, Yamada H (1992a) Nitrilase from Rhodococcus rhodochrous J1. Sequencing and overexpression of the gene and identification of an essential cysteine residue. J Biol Chem 267:20746–20751
Kobayashi M, Nagasawa T, Yamada H (1992b) Primary structure of an aliphatic nitrile-degrading enzyme, aliphatic nitrilase, from Rhodococcus rhodochrous K22 and expression of its gene and identification of its active site residue. Biochem 31:9000–9007
Komeda H, Hori Y, Kobayashi M, Shimizu S (1996) Transcriptional regulation of the Rhodococcus rhodochrous J1 nitA gene encoding a nitrilase. Proc Natl Acad Sci U S A 93:10572–10577
Kumar V, Kumar V, Thakur N, Bhalla TC (2015) Bench scale synthesis of p-hydroxybenzoic acid using whole-cell nitrilase of Gordonia terrae mutant E9. Bioprocess Biosyst Eng 38:1267–1279
Lathe WCIII, Snel B, Bork P (2000) Gene context conservation of a higher order than operons. Trends Biochem Sci 25:474–479
Lawrence JG (2003) Gene organization: selection, selfishness, and serendipity. Ann Rev Microbiol 57:419–440
Layh N, Parratt J, Willets A (1998) Characterization and partial purification of an enantioselective arylacetonitrilase from Pseudomonas fluorescens DSM 7155. J Mol Catal B Enzym 5:467–474
Lévy-Schil S, Soubrier F, Crutz-LeCoq A-M, Faucher D, Crouzet J, Pétré D (1995) Aliphatic nitrilase from a soil-isolated Comamonas testosteroni sp.: gene cloning and overexpression, purification and primary structure. Gene 161:15–20
Liu Z-Q, Dong L-Z, Cheng F, Xue Y-P, Wang Y-S, Ding J-V, Zheng Y-G, Shen Y-C (2011) Gene cloning, expression and characterization of a nitrilase from Alcaligenes faecalis. J Agric Food Chem 59:11560–11570
Luo H, Fan L, Chang Y, Ma J, Yu H, Shen Z (2010) Gene cloning, overexpression, and characterization of the nitrilase from Rhodococcus rhodochrous tg1-A6 in E. coli. Appl Biochem Biotechnol 160:393–400
Machingura M, Salomon E, Jez JM, Ebbs SD (2016) The β-cyanoalanine synthase pathway: beyond cyanide detoxification. Plant Cell Environ 39:2329–2341
Martínková L (2019) Nitrile metabolism in fungi: A review of its key enzymes nitrilases with focus on their biotechnological impact. Fungal Biol Rev. https://doi.org/10.1016/j.fbr.2018.11.002
Martínková L, Křen V (2010) Biotransformations with nitrilases. Curr Opin Chem Biol 14:130–137
Martínková L, Vejvoda V, Kaplan O, Kubáĉ D, Malandr A, Cantarella M, Bezouška K, Křen V (2009) Fungal nitrilases as biocatalysts: recent developments. Biotechnol Adv 27:661–670
Martínková L, Rucká L, Nešvera J, Pátek M (2017) Recent advances and challenges in the heterologous production of microbial nitrilases for biocatalytic applications. World J Microbiol Biotechnol 33:8
Matsumoto M, Hashimoto Y, Saitoh Y, Kumano T, Kobayashi M (2016) Development of nitrilase promoter-derived inducible vectors for Streptomyces. Biosci Biotechnol Biochem 80:1230–1237
Nagasawa T, Kobayashi M, Yamada H (1988) Optimum culture conditions for the production of benzonitrilase by Rhodococcus rhodochrous J1. Arch Microbiol 150:89–94
Nagasawa T, Mauger J, Yamada H (1990a) A novel nitrilase, arylacetonitrilase of Alcaligenes faecalis JM3. Eur J Biochem 194:765–772
Nagasawa T, Nakamura T, Yamada H (1990b) ε-Caprolactam, a new powerful inducer for the formation of Rhodococcus rhodochrous J1 nitrilase. Arch Microbiol 155:13–17
Nagasawa T, Wieser M, Nakamura T, Iwahara H, Yoshida T, Gekko K (2000) Nitrilase of Rhodococcus rhodochrous J1: conversion into the active form by subunit association. Eur J Biochem 267:138–144
Nageshwar YVD, Sheelu G, Shambhu RR, Muluka H, Mehdi N, Malik MS, Kamal A (2011) Optimization of nitrilase production from Alcaligenes faecalis MTCC 10757 (IICT-A43): effect of inducers on substrate specificity. Bioprocess Biosyst Eng 34:515–523
Niland P, Hühne R, Müller-Hill B (1996) How AraC interacts specifically with its target DNAs. J Mol Biol 264:667–674
Novikov AD, Riabchenko LE, Leonova TE, Larikova GA, Lavrov KV, Glinskii SA, Yanenko AS (2017) Bacterial strain Alcaligenes denitrificans C-32 containing two nitrilases with different substrate specificities. Appl Biochem Microbiol 53:786–791
Novo C, Farnaud S, Tata R, Clemente A, Brown PR (2002) Support for a three-dimensional structure predicting a Cys-Glu-Lys catalytic triad for Pseudomonas aeruginosa amidase comes from site-directed mutagenesis and mutations altering substrate specificity. Biochem J 365:731–738
O’Reilly C, Turner PD (2003) The nitrilase family of CN hydrolyzing enzymes—a comparative study. J Appl Microbiol 95:1161–1174
Pace HC, Brenner C (2001) The nitrilase superfamily: classification structure and function. Genome Biol 2:REVIEWS0001
Pai O, Banoth L, Ghosh S, Chisti Y, Banerjee UC (2014) Biotransformation of 3-cyanopyridine to nicotinic acid by free and immobilized cells of recombinant Escherichia coli. Process Biochem 49:655–659
Patel JM (2009) Biocatalytic synthesis of atorvastatin intermediates. J Mol Catal B Enzym 61:123–128
Petříčková A, Veselá AB, Kaplan O, Kubáč D, Uhnáková B, Malandra A, Felsberg J, Rinágelová A, Weyrauch P, Křen V, Bezouška K, Martínková L (2012) Purification and characterization of heterologously expressed nitrilases from filamentous fungi. Appl Microbiol Biotechnol 93:1553–1561
Piotrowski M (2008) Primary or secondary? Versatile nitrilases in plant metabolism. Phytochemistry 69:2655–2667
Podar M, Eads JR, Richardson TH (2005) Evolution of a microbial nitrilase gene family: a comparative and environmental genomics study. BMC Evol Biol 5:1–13
Prasad S, Misra A, Jangir VP, Awasthi A, Raj J, Bhalla TC (2007) A propionitrile induced nitrilase of Rhodococcus sp. NDB1165 and its application in nicotinic acid synthesis. World J Microbiol Biotechnol 23:345–353
Raczynska J, Vorgias C, Antranikian G, Rypniewski W (2010) Crystallographic structure of a thermoactive nitrilase. J Struct Biol 173:294–302
Rädisch R, Chmátal M, Rucká L, Novotný P, Petrásková L, Halada P, Kotik M, Pátek M, Martínková L (2018) Overproduction and characterization of the first enzyme of a new aldoxime dehydratase family in Bradyrhizobium sp. Int J Biol Macromol 115:746–753
Rezende RP, Teixeira DJC, Ferraz V, Linardi VR (2000) Metabolism of benzonitrile by Cryptococcus sp. UFMG-Y28. J Basic Microbiol 40:389–392
Rhee S, Martin RG, Rosner JL, Davies DR (1998) A novel DNA-binding motif in MarA: the first structure for an AraC family transcriptional regulator. Proc Natl Acad Sci U S A 95:10413–10418
Robertson DE, Chaplin JA, DeSantis G, Podar M, Madden M, Chi E, Richardson T, Milan A, Miller M, Weiner DP, Wong K, McQuaid J, Farwell B, Preston LA, Tan X, Snead MA, Keller M, Mathur E, Kretz PL, Burk MJ, Short JM (2004) Exploring nitrilase sequence space for enantioselective catalysis. Appl Environ Microbiol 70:2429–2436
Rogozin IB, Makarova KS, Murvaj J, Czabarka E, Wolf YI, Tatusov RL, Szekely LA, Koonin EV (2002) Connected gene neighborhoods in prokaryotic genomes. Nucleic Acids Res 30:2212–2223
Rosano GL, Ceccarelli EA (2014) Recombinant protein expression in Escherichia coli: advances and challenges. Front Microbiol 5:172
Rustler S, Stolz A (2007) Isolation and characterization of a nitrile hydrolysing acido tolerant black yeast-Exophiala oligosperma R1. Appl Microbiol Biotechnol 75:899–908
Sakashita T, Hashimoto Y, Oinuma K-I, Kobayashi M (2008) Transcriptional regulation of the nitrile hydratase gene cluster in Pseudomonas chlororaphis B23. J Bacteriol 190:4210–4217
Schϋller A, Slater AW, Norambuena T, Cifuentes JJ, Almonacid LI, Melo F (2012) Computer-based annotation of putative arac/xyls-family transcription factors of known structure but unknown function. J Biomed Biotechnol 2012:103132
Seffernick JL, Samanta SK, Louie TM, Wackett LP, Subramanian M (2009) Investigative mining of sequence data for novel enzymes: a case study with nitrilases. J Biotechnol 143:17–26
Sharma NN, Sharma M, Bhalla TC (2011) An improved nitrilase-mediated bioprocess for synthesis of nicotinic acid from 3-cyanopyridine with hyperinduced Nocardia globerula NHB-2. J Ind Microbiol Biotechnol 38:1235–1243
Sharma N, Thakur N, Raj T, Savitri, Bhalla TC (2017) Mining of microbial genomes for the novel sources of nitrilases. BioMed Res Int 2017:7039245
Sharma N, Verma R, Savitri, Bhalla TC (2018) Classifying nitrilases as aliphatic and aromatic using machine learning technique. 3 Biotech 8:68
Singh RV, Sharma H, Koul A, Babu V (2018) Exploring a broad spectrum nitrilase from moderately halophilic bacterium Halomonas sp. IIIMB2797 isolated from saline lake. J Basic Microbiol 58:876–874
Sosedov O, Stolz A (2014) Random mutagenesis of the arylacetonitrilase from Pseudomonas fluorescens EBC191 and identification of variants, which form increased amounts of mandeloamide from mandelonitrile. Appl Microbiol Biotechnol 98:1595–1607
Stalker DM, Malyj JD, McBride KE (1988) Purification and characterization of a nitrilase specific for the herbicide bromoxynil and corresponding nucleotide sequence analysis of the bxn gene. J Biol Chem 263:6310–6314
Stevenson DE, Feng R, Dumas F, Grolaeu D, Mihoc A, Storer AC (1992) Mechanistic and structural studies on Rhodococcus ATCC 39484 nitrilase. Biotechnol Appl Biochem 15:283–302
Sun J, Yu H, Chen J, Luo H, Shen Z (2016) Ammonium acrylate biomanufacturing by an engineered Rhodococcus ruber with nitrilase overexpression and double-knockout of nitrile hydratase and amidase. J Ind Microbiol Biotechnol 43:1631–1639
Taylor CM, Wang Q, Rosa BA, Huang SC-C, Powell K, Schedl T, Pearce EJ, Abubucker S, Mitreva M (2013) Discovery of anthelmintic drug targets and drugs using chokepoints in nematode metabolic pathways. PLoS Pathog 9(8):e1003505
Thakur N, Kumar V, Thakur S, Sharma N, Sheetal BT-C (2018) Biotransformation of 4-hydroxyphenylacetonitrile to 4-hydroxyphenylacetic acid using whole cell arylacetonitrilase of Alcaligenes faecalis MTCC 12629. Process Biochem 73:117–123
Thimann KV, Mahadevan S (1964) Nitrilase I. Occurrence, preparation and general properties of enzyme. Arch Biochem Biophys 105:133–141
Thuku RN, Weber BW, Varsani A, Sewell BT (2007) Post-translational cleavage of recombinantly expressed nitrilase from Rhodococcus rhodochrous J1 yields a stable, active helical form. FEBS J 274:2099–2108
Thuku RN, Brady D, Benedik MJ, Sewell BT (2009) Microbial nitrilases: versatile, spiral forming enzymes. J Appl Microbiol 106:703–727
Vejvoda V, Kaplan O, Klozová J, Masák J, Jirků ČA, Stloukal V, R. Martínková L. (2006) Mild hydrolysis of nitriles by Fusarium solani strain O1. Folia Microbiol 51:251–256
Vejvoda V, Kaplan O, Bezouška K, Pomach P, Šulc M, Cantarella M, Benada O, Uhnākovā B, Rināgelovā A, Wahl SL, Fischer L, Kŕen V, Martinkovā L (2008) Purification and characterization of a nitrilase from Fusarium solani O1. J Mol Catal B Enzym 50:99–106
Vejvoda V, Kubac D, Davidova A, Kaplan O, Sulc M, Sveda O, Chaloupkova R, Martinkova L (2010) Purification and characterization of nitrilase from Fusarium solani IMI196840. Process Biochem 45:1115–1120
Vergne-Vaxelaire C, Bordier F, Fossey A, Besnard-Gonnet M, Debard A, Mariage A, Pellouin V, Perret A, Petit J-L, Stam M, Salanoubat M, Weissenbach J, De Berardinis V, Zaparucha A (2013) Nitrilase activity screening on structurally diverse substrates: providing biocatalytic tools for organic synthesis. Adv Synth Catal 355:1763–1779
Veselá AB, Rucká L, Kaplan O, Pelantová H, Nešvera J, Pátek M, Martínková L (2016) Bringing nitrilase sequences from databases to life: the search for novel substrate specificities with a focus on dinitriles. Appl Microbiol Biotechnol 100:2193–2202
Wang H, Li G, Li M, Wei D, Wang X (2014) A novel nitrilase from Rhodobacter sphaeroides LHS-305: cloning, heterologous expression and biochemical characterization. World J Microbiol Biotechnol 30:245–252
Yamada H, Ryuno K, Nagasawa T, Enomoto K, Watanabe I (1986) Optimum culture conditions for production by Pseudomonas chlororaphis B23 of nitrile hydratase. Agric Biol Chem 50:2859–2865
Yamada H, Nagasawa T, Nakamura T (1991) Process for producing an elevated amount of nitrilase activity form a microbial strain of Rhodococcus. European Patent Office EP 0 444 640 B1. Filing date 27/2/1991
Yamamoto K, Komatsu K-I (1991) Purification and characterization of nitrilase responsible for the enantioselective hydrolysis from Acinetobacter sp. AK 226. Agric Biol Chem 55:1459–1466
Yamamoto K, Fijimatsu I, Komatsu K (1992) Purification and characterization of the nitrilase from Alcaligenes faecalis ATCC 8750 responsible for enantioselective hydrolysis of mandelonitrile. J Ferment Bioeng 73:425–430
Yang J, Zhang Y (2015) Protein structure and function prediction using i-TASSER. Curr Protoc Bioinformatics 52:5.8.1–5.815
Yang C, Wang X, Wei D (2011) A new nitrilase-producing strain named Rhodobacter sphaeroides LHS-305. Appl Biochem Biotechnol 165:1556–1567
Yusuf F, Chaubey A, Jamwal U, Parshad R (2013a) A new isolate from Fusarium proliferatum (AUF-2) for efficient nitrilase production. Appl Biochem Biotechnol 171:1022–1031
Yusuf F, Chaubey A, Raina A, Jamwal U, Parshad R (2013b) Enhancing nitrilase production from Fusarium proliferatum using response surface methodology. SpringerPlus 2:1–7
Zhang Z-J, Xy J-H, He Y-C, Ouyang L-M, Liy Y-Y (2011) Cloning and biochemical properties of a highly thermostable and enantioselective nitrilase from Alcaligenes sp. ECU0401 and its potential for (R)-(-)-mandelic acid production. Bioprocess Biosyst Eng 34:315–322
Zhang L, Yin B, Wang C, Jiang S, Wang H, Yuan YA, Wei D (2014) Structural insights into enzymatic activity and substrate specificity determination by a single amino acid in nitrilase from Synechocystis sp. PCC6803. J Struct Biol 188:93–101
Zheng YG, Chen J, Liu ZQ, Wu MH, Xing LY, Shen YC (2008) Isolation, identification and characterization of Bacillus subtilis ZJB-063, a versatile nitrile-converting bacterium. Appl Microbiol Biochem 77:985–993
Zhu D, Mukherjee C, Biehl ER, Hua L (2007) Discovery of a mandelonitrile hydrolase from Bradyrhizobium japonicum USDA110 by rational genome mining. J Biotechnol 129:645–650
Acknowledgments
We would like to thank the CSIR, the University of the Witwatersrand and DST for funding and resources needed to complete this work. Financial support from the Department of Science and Technology Biocatalysis Initiative (Grant 0175/2013) was greatly appreciated.
Funding
Department of Science and Technology Biocatalysis Initiative (Grant 0175/2013).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Ethical approval
This literature review does not contain any studies with human participants or animals by any of the authors.
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Chhiba-Govindjee, V.P., van der Westhuyzen, C.W., Bode, M.L. et al. Bacterial nitrilases and their regulation. Appl Microbiol Biotechnol 103, 4679–4692 (2019). https://doi.org/10.1007/s00253-019-09776-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-019-09776-1