Abstract
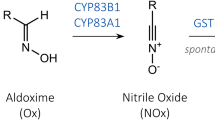
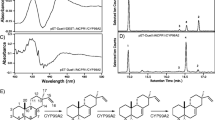
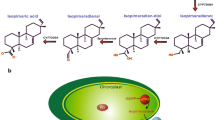
Cytochromes P450 (CYP) are attractive enzyme targets in biotechnology as they catalyze stereospecific C-hydroxylations of complex core skeletons at positions that typically are difficult to access by chemical synthesis. Membrane bound CYPs are involved in nearly all plant pathways leading to the formation of high-value compounds. In the present study, we systematically maximize the heterologous expression of six different plant-derived CYP genes in Escherichia coli, using a workflow based on C-terminal fusions to the green fluorescent protein. The six genes can be over-expressed in both K- and B-type E. coli strains using standard growth media. Furthermore, sequences encoding a small synthetic peptide and a small bacterial membrane anchor markedly enhance the expression of all six genes. For one of the CYPs, the length of the linker region between the predicted N-terminal transmembrane segment and the soluble domain is modified, in order to verify the importance of this region for enzymatic activity. The work describes how membrane bound CYPs are optimally produced in E. coli and thus adds this plant multi-membered key enzyme family to the toolbox for bacterial cell factory design.



Similar content being viewed by others
References
Bak S, Kahn RA, Nielsen HL, Moller BL, Halkier BA (1998a) Cloning of three A-type cytochromes P450, CYP71E1, CYP98, and CYP99 from Sorghum bicolor (L.) Moench by a PCR approach and identification by expression in Escherichia coli of CYP71E1 as a multifunctional cytochrome P450 in the biosynthesis of the cyanogenic glucoside dhurrin. Plant Mol Biol 36:393–405
Bak S, Kahn RA, Olsen CE, Halkier BA (1997) Cloning and expression in Escherichia coli of the obtusifoliol 14 alpha-demethylase of Sorghum bicolor (L.) Moench, a cytochrome P450 orthologous to the sterol 14 alpha-demethylases (CYP51) from fungi and mammals. Plant J 11:191–201
Bak S, Nielsen HL, Halkier BA (1998b) The presence of CYP79 homologues in glucosinolate-producing plants shows evolutionary conservation of the enzymes in the conversion of amino acid to aldoxime in the biosynthesis of cyanogenic glucosides and glucosinolates. Plant Mol Biol 38:725–734
Barnes HJ, Arlotto MP, Waterman MR (1991) Expression and enzymatic activity of recombinant cytochrome P450 17 alpha-hydroxylase in Escherichia coli. Proc Natl Acad Sci U S A 88:5597–5601
Biggs BW, Lim CG, Sagliani K, Shankar S, Stephanopoulos G, De Mey M, Ajikumar PK (2016) Overcoming heterologous protein interdependency to optimize P450-mediated Taxol precursor synthesis in Escherichia coli. Proc Natl Acad Sci U S A 113:3209–3214. doi:10.1073/pnas.1515826113
Chang MCY, Eachus RA, Trieu W, Ro D-K, Keasling JD (2007) Engineering Escherichia coli for production of functionalized terpenoids using plant P450s. Nat Chem Biol 3:274–277. doi:10.1038/nchembio875
Chau M, Jennewein S, Walker K, Croteau R (2004) Taxol biosynthesis: molecular cloning and of a cytochrome p450 characterization taxoid 7 beta-hydroxylase. Chemistry & Biology 11:663–672. doi:10.1016/j.chembiol.2004.02.025
Chen CD, Doray B, Kemper B (1998) A conserved proline-rich sequence between the N-terminal signal-anchor and catalytic domains is required for assembly of functional cytochrome P450 2C2. Arch Biochem Biophys 350:233–238. doi:10.1006/abbi.1997.0524
Costa S, Almeida A, Castro A, Domingues L (2014) Fusion tags for protein solubility, purification and immunogenicity in Escherichia coli: the novel Fh8 system. Front Microbiol 5:63. doi:10.3389/fmicb.2014.00063
Daley DO, Rapp M, Granseth E, Melen K, Drew D, Heijne von G (2005) Global topology analysis of the Escherichia coli inner membrane proteome. Science 308:1321–1323. doi:10.1126/science.1109730
Davies KM, Deroles SC (2014) Prospects for the use of plant cell cultures in food biotechnology. Curr Opin Biotechnol 26:133–140. doi:10.1016/j.copbio.2013.12.010
Denisov IG, Shih AY, Sligar SG (2012) Structural differences between soluble and membrane bound cytochrome P450s. J Inorg Biochem 108:150–158. doi:10.1016/j.jinorgbio.2011.11.026
Doray B, Chen CD, Kemper B (2001) N-terminal deletions and His-tag fusions dramatically affect expression of cytochrome p450 2C2 in bacteria. Arch Biochem Biophys 393:143–153. doi:10.1006/abbi.2001.2473
Drew D, Lerch M, Kunji E, Slotboom DJ, de Gier JW (2006) Optimization of membrane protein overexpression and purification using GFP fusions. Nat Methods 3:303–313. doi:10.1038/nmeth0406-303
Drew D, Slotboom D-J, Friso G, Reda T, Genevaux P, Rapp M, Meindl-Beinker NM, Lambert W, Lerch M, Daley DO, Van Wijk K-J, Hirst J, Kunji E, De Gier J-W (2005) A scalable, GFP-based pipeline for membrane protein overexpression screening and purification. Protein Sci 14:2011–2017. doi:10.1110/ps.051466205
Drew DE, Heijne von G, Nordlund P, de Gier JW (2001) Green fluorescent protein as an indicator to monitor membrane protein overexpression in Escherichia coli. FEBS Lett 507:220–224
Eugster HP, Bärtsch S, Würgler FE, Sengstag C (1992) Functional co-expression of human oxidoreductase and cytochrome P450 1A1 in Saccharomyces cerevisiae results in increased EROD activity. Biochem Biophys Res Commun 185:641–647
Gallage NJ, Møller BL (2015) Vanillin-bioconversion and bioengineering of the most popular plant flavor and its de novo biosynthesis in the vanilla orchid. Mol Plant 8:40–57. doi:10.1016/j.molp.2014.11.008
Hamberger B, Ohnishi T, Hamberger B, Séguin A, Bohlmann J (2011) Evolution of diterpene metabolism: Sitka spruce CYP720B4 catalyzes multiple oxidations in resin acid biosynthesis of conifer defense against insects. Plant Physiol 157:1677–1695. doi:10.1104/pp.111.185843
Hansen EH, Møller BL, Kock GR, Bünner CM, Kristensen C, Jensen OR, Okkels FT, Olsen CE, Motawia MS, Hansen J (2009) De novo biosynthesis of vanillin in fission yeast (Schizosaccharomyces pombe) and baker’s yeast (Saccharomyces cerevisiae). Appl Environ Microbiol 75:2765–2774. doi:10.1128/AEM.02681-08
Haudenschild C, Schalk M, Karp F, Croteau R (2000) Functional expression of regiospecific cytochrome P450 limonene hydroxylases from mint (Mentha spp.) in Escherichia coli and Saccharomyces cerevisiae. Arch Biochem Biophys 379:127–136. doi:10.1006/abbi.2000.1864
Hessa T, Kim H, Bihlmaier K, Lundin C, Boekel J, Andersson H, Nilsson I, White SH, Heijne von G (2005) Recognition of transmembrane helices by the endoplasmic reticulum translocon. Nature 433:377–381. doi:10.1038/nature03216
Jensen K, Osmani SA, Hamann T, Naur P, Møller BL (2011) Homology modeling of the three membrane proteins of the dhurrin metabolon: catalytic sites, membrane surface association and protein-protein interactions. Phytochemistry 72:2113–2123. doi:10.1016/j.phytochem.2011.05.001
Kawate T, Gouaux E (2006) Fluorescence-detection size-exclusion chromatography for precrystallization screening of integral membrane proteins. Structure 14:673–681. doi:10.1016/j.str.2006.01.013
Kim SH, Cavaleiro AM, Rennig M, Nørholm MHH (2016) SEVA linkers: a versatile and automatable DNA backbone exchange standard for synthetic biology. ACS Synth Biol 5:1177–1181. doi:10.1021/acssynbio.5b00257
Kitaoka N, Lu X, Yang B, Peters RJ (2015) The application of synthetic biology to elucidation of plant mono-, sesqui-, and diterpenoid metabolism. Plant Metabolism and Synthetic Biology 8:6–16. doi:10.1016/j.molp.2014.12.002
Koch BM, Sibbesen O, Halkier BA, Svendsen I, Moller BL (1995) The primary sequence of cytochrome P450tyr, the multifunctional N-hydroxylase catalyzing the conversion of L-tyrosine to p-hydroxyphenylacetaldehyde oxime in the biosynthesis of the cyanogenic glucoside dhurrin in Sorghum bicolor (L.) Moench. Arch Biochem Biophys 323:177–186
Laursen T, Møller BL, Bassard J-E (2015) Plasticity of specialized metabolism as mediated by dynamic metabolons. Trends Plant Sci 20:20–32. doi:10.1016/j.tplants.2014.11.002
Lee C, Kang HJ, Hjelm A, Qureshi AA, Nji E, Choudhury H, Beis K, De Gier J-W, Drew D (2014) MemStar: a one-shot Escherichia coli-based approach for high-level bacterial membrane protein production. FEBS Lett 588:3761–3769. doi:10.1016/j.febslet.2014.08.025
Leonard E, Koffas MAG (2007) Engineering of artificial plant cytochrome p450 enzymes for synthesis of isoflavones by Escherichia coli. Appl Environ Microbiol 73:7246–7251. doi:10.1128/AEM.01411-07
Miroux B, Walker JE (1996) Over-production of proteins in Escherichia coli: mutant hosts that allow synthesis of some membrane proteins and globular proteins at high levels. J Mol Biol 260:289–298. doi:10.1006/jmbi.1996.0399
Moller BL (2010) Dynamic metabolons. Science 330:1328–1329
Morant M, Bak S, Møller BL, Werck-Reichhart D (2003) Plant cytochromes P450: tools for pharmacology, plant protection and phytoremediation. Curr Opin Biotechnol 14:151–162
Morrone D, Chen X, Coates RM, Peters RJ (2010) Characterization of the kaurene oxidase CYP701A3, a multifunctional cytochrome P450 from gibberelin biosynthesis. Biochem J 431:337–347. doi:10.1042/BJ20100597
Møller BL (2014) Disruptive innovation: channeling photosynthetic electron flow into light-driven synthesis of high-value products. Synth Biol 1:330–359
Nelson DR (2011) Progress in tracing the evolutionary paths of cytochrome P450. Biochimica et Biophysica Acta (BBA) - Proteins and Proteomics 1814(1):14–18. doi:10.1016/j.bbapap.2010.08.008
Nour-Eldin HH, Hansen BG, Nørholm MHH, Jensen JK, Halkier BA (2006) Advancing uracil-excision based cloning towards an ideal technique for cloning PCR fragments. Nucleic Acids Res 34:e122–e122. doi:10.1093/nar/gkl635
Nørholm MHH (2010) A mutant Pfu DNA polymerase designed for advanced uracil-excision DNA engineering. BMC Biotechnol 10:21. doi:10.1186/1472-6750-10-21
Nørholm MHH, Toddo S, Virkki MTI, Light S, Heijne von G, Daley DO (2013) Improved production of membrane proteins in Escherichia coli by selective codon substitutions. FEBS Lett 587:2352–2358. doi:10.1016/j.febslet.2013.05.063
Paddon CJ, Westfall PJ, Pitera DJ, Benjamin K, Fisher K, McPhee D, Leavell MD, Tai A, Main A, Eng D, Polichuk DR, Teoh KH, Reed DW, Treynor T, Lenihan J, Fleck M, Bajad S, Dang G, Dengrove D, Diola D, Dorin G, Ellens KW, Fickes S, Galazzo J, Gaucher SP, Geistlinger T, Henry R, Hepp M, Horning T, Iqbal T, Jiang H, Kizer L, Lieu B, Melis D, Moss N, Regentin R, Secrest S, Tsuruta H, Vazquez R, Westblade LF, Xu L, Yu M, Zhang Y, Zhao L, Lievense J, Covello PS, Keasling JD, Reiling KK, Renninger NS, Newman JD (2013) High-level semi-synthetic production of the potent antimalarial artemisinin. Nature:496–528. doi:10.1038/nature12051
Pritchard MP, Ossetian R, Li DN, Henderson CJ, Burchell B, Wolf CR, Friedberg T (1997) A general strategy for the expression of recombinant human cytochrome P450s in Escherichia coli using bacterial signal peptides: expression of CYP3A4, CYP2A6, and CYP2E1. Arch Biochem Biophys 345:342–354. doi:10.1006/abbi.1997.0265
Qi X, Bakht S, Qin B, Leggett M, Hemmings A, Mellon F, Eagles J, Werck-Reichhart D, Schaller H, Lesot A, Melton R, Osbourn A (2006) A different function for a member of an ancient and highly conserved cytochrome P450 family: from essential sterols to plant defense. Proc Natl Acad Sci U S A 103:18848–18853. doi:10.1073/pnas.0607849103
Silva-Rocha R, Martínez-García E, Calles B, Chavarría M, Arce-Rodríguez A, de Las Heras A, Páez-Espino AD, Durante-Rodríguez G, Kim J, Nikel PI, Platero R, De Lorenzo V (2013) The Standard European Vector Architecture (SEVA): a coherent platform for the analysis and deployment of complex prokaryotic phenotypes. Nucleic Acids Res 41:D666–D675. doi:10.1093/nar/gks1119
Sonnhammer EL, Heijne von G, Krogh A (1998) A hidden Markov model for predicting transmembrane helices in protein sequences. Proc Int Conf Intell Syst Mol Biol 6:175–182
Sonoda Y, Cameron A, Newstead S, Omote H, Moriyama Y, Kasahara M, Iwata S, Drew D (2010) Tricks of the trade used to accelerate high-resolution structure determination of membrane proteins. FEBS Lett 12:2539–2547. doi:10.1016/j.febslet.2010.04.015
Sonoda Y, Newstead S, Hu N-J, Alguel Y, Nji E, Beis K, Yashiro S, Lee C, Leung J, Cameron AD, Byrne B, Iwata S, Drew D (2011) Benchmarking membrane protein detergent stability for improving throughput of high-resolution X-ray structures. Structure 19:17–25. doi:10.1016/j.str.2010.12.001
Studier FW (1991) Use of bacteriophage T7 lysozyme to improve an inducible T7 expression system. J Mol Biol 219:37–44
Studier FW (2005) Protein production by auto-induction in high-density shaking cultures. Protein Expr Purif 41:207–234. doi:10.1016/j.pep.2005.01.016
Sudhamsu J, Kabir M, Airola MV, Patel BA, Yeh S-R, Rousseau DL, Crane BR (2010) Co-expression of ferrochelatase allows for complete heme incorporation into recombinant proteins produced in E. coli. Protein Expr Purif 73:78–82. doi:10.1016/j.pep.2010.03.010
Søgaard KM, Nørholm MHH (2016) Side effects of extra tRNA supplied in a typical bacterial protein production scenario. Protein Sci 25:2102–2108. doi:10.1002/pro.3011
Wadsäter M, Laursen T, Singha A, Hatzakis NS, Stamou D, Barker R, Mortensen K, Feidenhans’l R, Møller BL, Cárdenas M (2012) Monitoring shifts in the conformation equilibrium of the membrane protein cytochrome P450 reductase (POR) in nanodiscs. J Biol Chem 287:34596–34603. doi:10.1074/jbc.M112.400085
Wagner S, Klepsch MM, Schlegel S, Appel A, Draheim R, Tarry M, Hogbom M, Van Wijk KJ, Slotboom DJ, Persson JO, de Gier JW (2008) Tuning Escherichia coli for membrane protein overexpression. Proc Natl Acad Sci U S A 105:14371–14376. doi:10.1073/pnas.0804090105
Williams PA, Cosme J, Sridhar V, Johnson EF, McRee DE (2000) Mammalian microsomal cytochrome P450 monooxygenase: structural adaptations for membrane binding and functional diversity. Mol Cell 5:121–131
Zelasko S, Palaria A, Das A (2013) Optimizations to achieve high-level expression of cytochrome P450 proteins using Escherichia coli expression systems. Protein Expr Purif 92:77–87. doi:10.1016/j.pep.2013.07.017
Zhang H, Im S-C, Waskell L (2007) Cytochrome b(5) increases the rate of product formation by cytochrome p450 2B4 and competes with cytochrome p450 reductase for a binding site on cytochrome p450 2B4. J Biol Chem 282:29766–29776. doi:10.1074/jbc.M703845200
Acknowledgements
We thank Victor de Lorenzo and the members of his laboratory for generously providing the pSEVA collection. We thank Tomas Laursen, Peter Naur, Søren Bak, Björn Hamberger, Johan Andersen-Ranberg, and Britta Hamberger for advice on CYPs and reductases. We thank David Drew, Daniel Daley, and Jan. Willem de Gier for discussions on E. coli gene expression and the GFP-based expression platform. SS is the recipient of VILLUM Foundation’s Young Investigator Programme grant VKR023128. This work was supported by the Novo Nordisk Foundation, from the VILLUM research center of excellence “Plant Plasticity,” from the UCPH Excellence Program for Interdisciplinary Research to Center of Synthetic Biology “bioSYNergy,” and by a European Research Council Advanced Grant Project No. 323034: LightdrivenP450s.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Ulla Christensen and Dario Vazquez-Albacete contributed equally to this work.
Electronic supplementary material
ESM 1
(PDF 2837 kb)
Rights and permissions
About this article
Cite this article
Christensen, U., Vazquez-Albacete, D., Søgaard, K.M. et al. De-bugging and maximizing plant cytochrome P450 production in Escherichia coli with C-terminal GFP fusions. Appl Microbiol Biotechnol 101, 4103–4113 (2017). https://doi.org/10.1007/s00253-016-8076-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-016-8076-5