Abstract
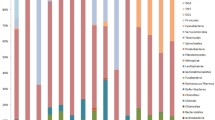
Coral-associated bacteria are critical for the well-being of their host and may play essential roles during ontogeny, as suggested by the vertical transmission of some bacteria in brooding corals. Bacterial acquisition patterns in broadcast spawners remain uncertain, as 16S rRNA gene metabarcoding of coral early life stages suggests the presence of bacterial communities, which have not been detected by microscopic examinations. Here, we combined 16S rRNA gene metabarcoding with fluorescence in situ hybridization (FISH) microscopy to analyze bacterial assemblages in Acropora tenuis egg–sperm bundles, embryos, and larvae following a spawning event. Metabarcoding results indicated that A. tenuis offspring ≤ 4-day-old were associated with diverse and dynamic bacterial microbiomes, dominated by Rhodobacteraceae, Alteromonadaceae, and Oceanospirillaceae. While FISH analyses confirmed the lack of internalized bacteria in A. tenuis offspring, metabarcoding showed that even the earliest life stages examined (egg–sperm bundles and two-cell stages) were associated with a diverse bacterial community, suggesting the bacteria were confined to the mucus layer. These results can be explained by vertical transmission of certain taxa (mainly Endozoicomonas) in the mucus surrounding the gametes within bundles, or by horizontal bacterial transmission through the release of bacteria by spawning adults into the water column.







Similar content being viewed by others
Availability of Data and Material
The sequence data generated and analyzed in this study are available at NCBI under http://www.ncbi.nlm.nih.gov/bioproject/517286, BioProject accession PRJNA517286.
References
Amann RI, Binder BJ, Olson RJ, Chisholm SW, Devereux R, Stahl DA (1990) Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl. Environ. Microbiol. 56:1919–1925
Anderson MJ (2001) A new method for non-parametric multivariate analysis of variance. Austral Ecology 26:32–46
Anderson MJ (2006) Distance-based tests for homogeneity of multivariate dispersions. Biometrics 62:245–253
Anderson MJ, Ellingsen KE, McArdle BH (2006) Multivariate dispersion as a measure of beta diversity. Ecol. Lett. 9:683–693
Andersson AF, Lindberg M, Jakobsson H, Bäckhed F, Nyrén P, Engstrand L (2008) Comparative analysis of human gut microbiota by barcoded pyrosequencing. PLoS One 3:e2836
Apprill A, Marlow HQ, Martindale MQ, Rappe MS (2009) The onset of microbial associations in the coral Pocillopora meandrina. ISME J 3:685–699
Apprill A, Marlow HQ, Martindale MQ, Rappe MS (2012) Specificity of associations between bacteria and the coral Pocillopora meandrina during early development. Appl. Environ. Microbiol. 78:7467–7475
Baird AH, Guest JR, Willis BL (2009) Systematic and biogeographical patterns in the reproductive biology of Scleractinian corals. Annu. Rev. Ecol. Evol. Syst. 40:551–571
Baker AC, Starger CJ, McClanahan TR, Glynn PW (2004) Corals’ adaptive response to climate change: shifting to new algal symbionts may safeguard devastated reefs from extinction. Nature 430:741
Bayer T, Neave MJ, Alsheikh-Hussain A, Adanda M, Yum LK, Mincer T, Hughen K, Apprill A, Voolstra CR (2013) The microbiome of the Red Sea coral Stylophora pistillata is dominated by tissue-associated Endozoicomonas bacteria. Appl. Environ. Microbiol. 79:4759–4762
Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. 57:289–300
Berkelmans R, van Oppen MJH (2006) The role of zooxanthellae in the thermal tolerance of corals: a ‘nugget of hope’ for coral reefs in an era of climate change. Proc Biol Sci / R Soc 273:2305–2312
Bernasconi R, Stat M, Koenders A, Paparini A, Bunce M, Huggett MJ (2019) Establishment of coral-bacteria symbioses reveal changes in the core bacterial community with host ontogeny. Front. Microbiol. 10:1529
Blackall LL, Wilson B, van Oppen MJ (2015) Coral—the world’s most diverse symbiotic ecosystem. Mol. Ecol. 24:5330–5347
Bokulich NA, Subramanian S, Faith JJ, Gevers D, Gordon JI, Knight R, Mills DA, Caporaso JG (2013) Quality-filtering vastly improves diversity estimates from Illumina amplicon sequencing. Nat. Methods 10:57–59
Boulotte NM, Dalton SJ, Carroll AG, Harrison PL, Putnam HM, Peplow LM, van Oppen MJ (2016) Exploring the Symbiodinium rare biosphere provides evidence for symbiont switching in reef-building corals. ISME J 10:2693–2701
Bourne DG, Morrow KM, Webster NS (2016) Insights into the coral microbiome: underpinning the health and resilience of reef ecosystems. Annu. Rev. Microbiol. 70:317–340
Bruno JF, Selig ER (2007) Regional decline of coral cover in the Indo-Pacific: timing, extent, and subregional comparisons. PLoS One 2:1–8
Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJ, Holmes SP (2016) DADA2: high-resolution sample inference from Illumina amplicon data. Nat. Methods 13:581–583
Callahan BJ, McMurdie PJ, Holmes SP (2017) Exact sequence variants should replace operational taxonomic units in marker-gene data analysis. The ISME journal 11:2639–2643
Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Gonzalez Pena A, Goodrich JK, Gordon JI, Huttley GA, Kelley S, Knights D, Koening JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R (2010) QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7:335–336
Ceh J, Raina JB, Soo RM, van Keulen M, Bourne DG (2012) Coral-bacterial communities before and after a coral mass spawning event on Ningaloo Reef. PLoS One 7:e36920
Ceh J, van Keulen M, Bourne DG (2013a) Intergenerational transfer of specific bacteria in corals and possible implications for offspring fitness. Microb. Ecol. 65:227–231
Ceh J, Kilburn MR, Cliff JB, Raina J-B, van Keulen M, Bourne DG (2013b) Nutrient cycling in early coral life stages: Pocillopora damicornis larvae provide their algal symbiont (Symbiodinium) with nitrogen acquired from bacterial associates. Ecol Evol 3:2393–2400
Chiu H-H, Mette A, Shiua J-H, Tang SL (2012) Bacterial distribution in the epidermis and mucus of the coral Euphyllia glabrescens by CARD-FISH. Zool Stud. 51:1332–1342
Courtesy of Integration and Application Network , IAN Symbol Libraries, ian.umces.edu/symbols/ Accessed 2018
Damjanovic K, van Oppen MJH, Menéndez P, Blackall LL, (2019) Experimental inoculation of coral recruits with marine bacteria indicates scope for microbiome manipulation in Acropora tenuis and Platygyra daedalea. Front Microbiol 10:1702
Daniels CA, Zeifman A, Heym K, Ritchie KB, Watson CA, Berzins I, Breitbart M (2011) Spatial heterogeneity of bacterial communities in the mucus of Montastraea annularis. Mar Ecol Prog Ser 426:29–40
De Cáceres M, Legendre P (2009) Associations between species and groups of sites: indices and statistical inference. Ecology 90:3566–3574
Durante MK, Baums IB, Williams DE, Vohsen S, Kemp DW (2019) What drives phenotypic divergence among coral clonemates of Acropora palmata? Mol. Ecol. 28(13):3208–3322
Epstein H, Torda G, Munday PL, van Oppen MJH (2019) Parental and early life stage environments drive establishment of bacterial and dinoflagellate communities in a common coral. ISME J 13:1635–1638
Frade PR, Roll K, Bergauer K, Herndl GJ (2016) Archaeal and bacterial communities associated with the surface mucus of Caribbean corals differ in their degree of host specificity and community turnover over reefs. PLoS One 11:e0144702
Harrison PL (2011) Sexual reproduction of scleractinian corals. In: Dubinsky Z, Stambler N (eds) Coral reefs: an ecosystem in transition. Springer, pp 59–85
Harrison PL, Booth DJ (2007) Coral reefs: naturally dynamic and increasingly disturbed ecosystems. In: Connell SD, Gillanders BM (eds) Marine ecology. Oxford University Press, Melbourne, pp 316–377
Herve M (2018) RVAideMemoire: testing and plotting procedures for biostatistics. R package version 0.9–69
Hoegh-Guldberg O (2011) The impact of climate change on coral reef ecosystems. In: Dubinsky Z, Stambler N (eds) Coral reefs: an ecosystem in transition. Springer, pp 391–403
Hoegh-Guldberg O, Kennedy EV, Beyer HL, McClennen C, Possingham HP (2018) Securing a long-term future for coral reefs. Trends Ecol Evol 33(12):936–944
Hothorn T, Bretz F, Westfall P (2008) Simultaneous inference in general parametric models. Biom. J. 50:346–363
Hugenholtz P, Tyson GW, Blackall LL (2002) Design and evaluation of 16S rRNA-targeted oligonucleotide probes for fluorescence in situ hybridization. Methods Mol. Biol. 179:29–42
Hughes JB, Hellmann JJ (2005) The application of rarefaction techniques to molecular inventories of microbial diversity. Methods Enzymol. 397:292–308
Hughes TP, Anderson KD, Connolly SR, Heron SF, Kerry JT, Lough JM, Baird AH, Baum JK, Berumen ML, Bridge TC, Claar DC, Eakin CM, Hobbs J-PA, Hoey AS, Hoogenboom MO, Lowe RJ, McCulloch MT, Pandolfi JM, Pratchett M, Schoepf V, Torda G, Wilson SK (2018) Spatial and temporal patterns of mass bleaching of corals in the Anthropocene. Science 359:80–83
Johansson ME, Hansson GC (2012) Preservation of mucus in histological sections, immunostaining of mucins in fixed tissue, and localization of bacteria with FISH. Methods Mol Biol 842:229–235
Lande R (1996) Statistics and partitioning of species diversity, and similarity among multiple communities. Oikos 76:5–13
Lee MD, Walworth NG, Sylvan JB, Edwards KJ, Orcutt BN (2015) Microbial communities on seafloor basalts at Dorado Outcrop reflect level of alteration and highlight global lithic clades. Front Microbiol 6:1470
Legendre P, Legendre L (1998) Numerical ecology (developments in environmental modelling). Elsevier Science
Leite DCA, Leao P, Garrido AG, Lins U, dos Santos HF, Pires DO, Castro CB, van Elsas JD, Zilberberg C, Rosado AS, Peixoto RS (2017) Broadcast spawning coral Mussismilia hispida can vertically transfer its associated bacterial core. Front Microbiol 8:1–12
Lema KA, Bourne DG, Willis BL (2014) Onset and establishment of diazotrophs and other bacterial associates in the early life history stages of the coral Acropora millepora. Mol. Ecol. 23:4682–4695
Lema KA, Clode PL, Kilburn MR, Thornton R, Willis BL, Bourne DG (2016) Imaging the uptake of nitrogen-fixing bacteria into larvae of the coral Acropora millepora. ISME J 10:1804–1808
Lesser MP, Falcón LI, Rodríguez-Román A, Enríquez S, Hoegh-Guldberg O, Iglesias-Prieto R (2007) Nitrogen fixation by symbiotic cyanobacteria provides a source of nitrogen for the scleractinian coral Montastraea cavernosa. Mar Ecol Prog Ser 346:143–152
Littman RA, Willis BL, Bourne DG (2009) Bacterial communities of juvenile corals infected with different Symbiodinium (dinoflagellate) clades. Mar Ecol Prog Ser 389:45–59
Love MI, Huber W, Anders S (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15:550
McDonald D, Clemente JC, Kuczynski J, Rideout JR, Stombaugh J, Wendel D, Wilke A, Huse S, Hufnagle J, Meyer F, Knight R, Caporaso JG (2012) The biological observation matrix (BIOM) format or: how I learned to stop worrying and love the ome-ome. GigaScience 1:1–6
McMurdie PJ, Holmes S (2013) phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One 8:e61217
Muscatine L, Porter JW (1977) Reef corals—mutualistic symbioses adapted to nutrient-poor environments. Bioscience 27:454–460
Nissimov J, Rosenberg E, Munn CB (2009) Antimicrobial properties of resident coral mucus bacteria of Oculina patagonica. FEMS Microbiol Lett 292:210–215
Oksanen J, Blanchet FG, Friendly M, Kindt R, Legendre P, McGlinn D, Minchin PR, O'Hara RB, Simpson CL, Solymos P, Henry M, Stevens H, Szoecs E, Wagner H (2016) Vegan: community ecology package. R package version 2.4–1
Padilla-Gamiño JL, Weatherby TM, Waller RG, Gates RD (2010) Formation and structural organization of the egg–sperm bundle of the scleractinian coral Montipora capitata. Coral Reefs 30:371–380
Pollock FJ, McMinds R, Smith S, Bourne DG, Willis BL, Medina M, Vega Thurber R, Zaneveld JR (2018) Coral-associated bacteria demonstrate phylosymbiosis and cophylogeny. Nat. Commun. 9:1–13
Porter TM, Hajibabaei M (2018) Scaling up: a guide to high-throughput genomic approaches for biodiversity analysis. Mol. Ecol. 27:313–338
QIIME 2 Development Team, QIIME 2, https://docs.qiime2.org Accessed 2017a
QIIME 2 Development Team, q2-demux, https://github.com/qiime2/q2-demux Accessed 2017b
QIIME 2 Development Team, q2-feature-classifier, https://github.com/qiime2/q2-feature-classifier Accessed 2017c
QIIME 2 Development Team, q2-taxa, https://github.com/qiime2/q2-taxa Accessed 2017d
Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Glockner FO (2013) The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41:D590–D596
R Core Team, R: a language and environment for statistical computing. http://www.R-project.org. Accessed 2018
Raina JB, Tapiolas D, Willis BL, Bourne DG (2009) Coral-associated bacteria and their role in the biogeochemical cycling of sulfur. Appl Environ Microbiol 75:3492–3501
Raina JB, Clode PL, Cheong S, Bougoure J, Kilburn MR, Reeder A, Foret S, Stat M, Beltran V, Thomas-Hall P, Tapiolas D, Motti CM, Gong B, Pernice M, Marjo CE, Seymour JR, Willis BL, Bourne DG (2017) Subcellular tracking reveals the location of dimethylsulfoniopropionate in microalgae and visualises its uptake by marine bacteria. eLIFE 6:e23008
Reshef L, Koren O, Loya Y, Zilber-Rosenberg I, Rosenberg E (2006) The coral probiotic hypothesis. Environ. Microbiol. 8:2068–2073
Ricardo GF, Jones RJ, Negri AP, Stocker R (2016a) That sinking feeling: suspended sediments can prevent the ascent of coral egg bundles. Sci. Rep. 6:21567
Ricardo GF, Jones RJ, Clode PL, Negri AP (2016b) Mucous secretion and cilia beating defend developing coral larvae from suspended sediments. PLoS One 11:e0162743
Ritchie KB (2006) Regulation of microbial populations by coral surface mucus and mucus-associated bacteria. Mar Ecol Prog Ser 322:1–14
Rohwer F, Seguritan V, Azam F, Knowlton N (2002) Diversity and distribution of coral-associated bacteria. Mar Ecol Prog Ser 243:1–10
Rosenberg E, Koren O, Reshef L, Efrony R, Zilber-Rosenberg I (2007) The role of microorganisms in coral health, disease and evolution. Nat Rev Microbiol 5:355–362
Röthig T, Ochsenkuhn MA, Roik A, van der Merwe R, Voolstra CR (2016) Long-term salinity tolerance is accompanied by major restructuring of the coral bacterial microbiome. Mol Ecol 25:1308–1323
Salter SS, Cox MJ, Turek EM, Calus ST, Cookson WO, Moffatt MF, Turner P, Parkhill J, Loman NJ, Walker AW (2014) Reagent and laboratory contamination can critically impact sequence-based microbiome analyses. BMC Biol. 12:1–12
Sharp KH, Ritchie KB, Schupp PJ, Ritson-Williams R, Paul VJ (2010) Bacterial acquisition in juveniles of several broadcast spawning coral species. PLoS One 5:e10898
Sharp KH, Distel D, Paul VJ (2012) Diversity and dynamics of bacterial communities in early life stages of the Caribbean coral Porites astreoides. ISME J 6:790–801
Shnit-Orland M, Kushmaro A (2009) Coral mucus-associated bacteria: a possible first line of defense. FEMS Microbiol Ecol 67:371–380
Sunagawa S, Woodley CM, Medina M (2010) Threatened corals provide underexplored microbial habitats. PLoS One 5:1–7
Sweet MJ, Croquer A, Bythell JC (2010) Bacterial assemblages differ between compartments within the coral holobiont. Coral Reefs 30:39–52
van Oppen MJH, Oliver JK, Putnam HM, Gates RD (2015) Building coral reef resilience through assisted evolution. Proc Natl Acad Sci 112:2307–2313
Wada N, Pollock FJ, Willis BL, Ainsworth T, Mano N, Bourne DG (2016) In situ visualization of bacterial populations in coral tissues: pitfalls and solutions. PeerJ 4:e2424
Wallner G, Amann R, Beisker W (1993) Optimizing fluorescent in situ hybridization with rRNA-targeted oligonucleotide probes for flow cytometric identification of microorganisms. Cytometry 14:136–143
Webster NS, Reusch T (2017) Microbial contributions to the persistence of coral reefs. ISME J:1–8
Weiss S, Xu ZZ, Peddada S, Amir A, Bittinger K, Gonzalez A, Lozupone C, Zaneveld JR, Vazquez-Baeza Y, Birmingham A, Hyde ER, Knight R (2017) Normalization and microbial differential abundance strategies depend upon data characteristics. Microbiome 5:27
Wickham H (2009) ggplot2: elegant graphics for data analysis. Springer-Verlag, New York
Wickham H (2017) tidyverse: easily install and load the ‘Tidyverse’. R package version 1.2.1
Zhang Y, Ling J, Yang Q, Wen C, Yan Q, Sun H, Van Nostrand JD, Shi Z, Zhou J, Dong J (2015) The functional gene composition and metabolic potential of coral-associated microbial communities. Sci. Rep. 5:16191
Zhou G, Cai L, Yuan T, Tian R, Tong H, Zhang W, Jiang L, Guo M, Liu S, Qian PY, Huang H (2017) Microbiome dynamics in early life stages of the scleractinian coral Acropora gemmifera in response to elevated pCO2. Environ Microbiol 19:3342–3352
Ziegler M, Seneca FO, Yum LK, Palumbi SR, Voolstra CR (2017) Bacterial community dynamics are linked to patterns of coral heat tolerance. Nat Commun 8:14213
Acknowledgments
The authors thank the team of the National Sea Simulator at AIMS for technical support in conducting the experiment. KD is very grateful to Dr. Naohisa Wada and Dr. Nicole Webster for extensive discussions and insights about FISH on coral samples, and also thanks Dr. David Bourne, Dr. Koty Sharp, and Dr. Kim Lema for their advice on the topic. We are grateful to Dr. Gabriela Segal for her instructions regarding the use of the confocal microscope and Zeiss software. Many thanks to Dr. Andrew Negri, Leela Chakravarti, and Lesa Peplow for their assistance during spawning, to Ashley Dungan for logistical help, and to Marie Roman for the images of adult coral and egg–sperm bundles used in Fig. 5.
Funding
This work was funded by an Australian Research Council grant (DP160101468) to van Oppen and Blackall, the Australian Institute of Marine Science (AIMS) and a Holsworth Wildlife Research Endowment (Equity Trustees Charitable Foundation & the Ecological Society of Australia). KD is the recipient of an International Postgraduate Research Scholarship and Australian Post-graduate Award (University of Melbourne). MvO acknowledges Australian Research Council Laureate Fellowship FL180100036.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare that they have no competing interests.
Rights and permissions
About this article
Cite this article
Damjanovic, K., Menéndez, P., Blackall, L.L. et al. Early Life Stages of a Common Broadcast Spawning Coral Associate with Specific Bacterial Communities Despite Lack of Internalized Bacteria. Microb Ecol 79, 706–719 (2020). https://doi.org/10.1007/s00248-019-01428-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00248-019-01428-1