Abstract
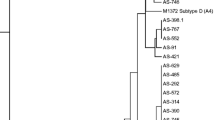
Transmissible hypovirulence associated with Cryphonectria hypovirus 1 (CHV1) has been used for biological control of chestnut blight, devastating disease of chestnut caused by the fungus Cryphonectria parasitica. The main aims of this study were to provide molecular characterization of CHV1 from Croatia and Slovenia and to reveal its genetic variability, phylogeny, and diversification of populations. Fifty-one CHV1 haplotypes were detected among 54 partially sequenced CHV1 isolates, all belonging to Italian subtype (I). Diversity was mainly generated by point mutations while evidence of recombination was not found. The level of conservation over analyzed parts of ORF-A proteins p29 and p40 varied, but functional sites were highly conserved. Phylogenetic analysis revealed close relatedness and intermixing of Croatian and Slovenian CHV1 populations. Our CHV1 isolates were also related to Swiss and Bosnian hypoviruses supporting previously suggested course of CHV1 invasion in Europe. Overall, this study indicates that phylogeny of CHV1 subtype I in Europe is complex and characterized with frequent point mutations resulting in many closely related variants of the virus. Possible association between variations within CHV1 ORF-A and growth of the hypovirulent fungal isolates is tested and presented.






Similar content being viewed by others
References
Pearson MN, Beever RE, Boine B, Arthur K (2009) Mycoviruses of filamentous fungi and their relevance to plant pathology. Mol Plant Pathol 10:115–128. https://doi.org/10.1111/j.1364-3703.2008.00503.x
Heiniger U, Rigling D (1994) Biological control of chestnut blight in Europe. Annu Rev Phytopathol 32:581–599. https://doi.org/10.1146/annurev.py.32.090194.003053
Milgroom MG, Cortesi P (2004) Biological control of chestnut blight with hypovirulence: a critical analysis. Annu Rev Phytopathol 42:311–338. https://doi.org/10.1146/annurev.phyto.42.040803.140325
Prospero S, Conedera M, Heiniger U, Rigling D (2006) Saprophytic activity and sporulation of Cryphonectria parasitica on dead chestnut wood in forests with naturally established hypovirulence. Phytopathology 96:1337–1344. https://doi.org/10.1094/PHYTO-96-1337
Nuss DL (2005) Hypovirulence: mycoviruses at the fungal-plant interface. Nat Rev Microbiol 3:632–642. https://doi.org/10.1038/nrmicro1206
Craven MG, Pawlyk MD, Choi GH, Nuss DL (1993) Papain-like protease p29 as a symptom determinant encoded by a hypovirulence-associated virus of the chestnut blight fungus. J Virol 67:6513–6521 PMID: 8411354
Suzuki N, Maruyama K, Moriyama M, Nuss DL (2003) Hypovirus papain-like protease p29 functions in trans to enhance viral double-stranded RNA accumulation and vertical transmission. J Virol 77:11697–11707. https://doi.org/10.1128/JVI.77.21.11697-11707.2003
Suzuki N, Nuss DL (2002) The contribution of p40 to hypovirus-mediated modulation of fungal host phenotype and viral RNA accumulation. J Virol 76:7747–7759. https://doi.org/10.1128/JVI.76.15.7747-7759.2002
Gobbin D, Hoegger PJ, Heiniger U, Rigling D (2003) Sequence variation and evolution of Cryphonectria hypovirus 1 (CHV1) in Europe. Virus Res 97:39–46. https://doi.org/10.1016/S0168-1702(03)00220-X
Feau N, Dutech C, Brusini J, Rigling D, Robin C (2014) Multiple introductions and recombination in Cryphonectria hypovirus 1: perspective for a sustainable biological control of chestnut blight. Evol Appl 7:580–596. https://doi.org/10.1111/eva.12157
Sotirovski K, Milgroom MG, Rigling D, Heiniger U (2006) Occurrence of Cryphonectria hypovirus1 in the chestnut blight fungus in Macedonia. Forest Pathol 36:136–143. https://doi.org/10.1111/j.1439-0329.2006.00443.x
Robin C, Lanz S, Soutrenon A, Rigling D (2010) Dominance of natural over released biological control agents of the chestnut blight fungus Cryphonectria parasitica in southeastern France is associated with fitness-related traits. Biol Control 53:55–61. https://doi.org/10.1016/j.biocontrol.2009.10.013
Krstin L, Novak-Agaba S, Rigling D, Ćurkovic-Perica M (2011) Diversity of vegetative compatibility types and mating types of Cryphonectria parasitica in Slovenia and occurrence of associated Cryphonectria hypovirus 1. Plant Pathol 60:752–761. https://doi.org/10.1111/j.1365-3059.2011.02438.x
Zamora P, Martin AB, Rigling D, Diez JJ (2012) Diversity of Cryphonectria parasitica in western Spain and identification of hypovirus-infected isolates. Forest Pathol 42:412–419. https://doi.org/10.1111/j.1439-0329.2012.00775.x
Akilli S, Serce CU, Katircioglu YZ, Maden S, Rigling D (2013) Characterization of hypovirulent isolates of the chestnut blight fungus, Cryphonectria parasitica from the Marmara and Black Sea regions of Turkey. Eur J Plant Pathol 135:323–334. https://doi.org/10.1007/s10658-012-0089-z
Peters FS, Bußkamp J, Prospero S, Rigling D, Metzler B (2014) Genetic diversification of the chestnut blight fungus Cryphonectria parasitica and its associated hypovirus in Germany. Fungal Biol 118:193–210. https://doi.org/10.1016/j.funbio.2013.11.009
Roderick GK, Hufbauer R, Navajas M (2012) Evolution and biological control. Evol Appl 5:419–423. https://doi.org/10.1111/j.1752-4571.2012.00281.x
Mlinarec J, Nuskern L, Ježić M, Rigling D, Ćurković-Perica M (2018) Molecular evolution and invasion pattern of Cryphonectria hypovirus 1 in Europe: Mutation rate, and selection pressure differ between genome domains. Virology 15:156–164. https://doi.org/10.1016/j.virol.2017.11.011
Bryner SF, Rigling D, Brunner PC (2012) Invasion history and demographic pattern of Cryphonectria hypovirus 1 across European populations of the chestnut blight fungus. Ecol Evol 2:3227–3241. https://doi.org/10.1002/ece3.429
Krstin L, Novak-Agbaba S, Rigling D, Krajačić M, Ćurković-Perica M (2008) Chestnut blight fungus in Croatia: diversity of vegetative compatibility types, mating types and genetic variability of associated Cryphonectria hypovirus 1. Plant Pathol 57:1086–1096. https://doi.org/10.1111/j.1365-3059.2008.01905.x
Krstin L, Krajačić M, Ćurković-Perica M, Novak-Agbaba S, Rigling D (2009) Hypovirus-infected strains of the fungus Cryphonectria parasitica in the central part of Croatia. Acta Hortic 815:283–287. https://doi.org/10.17660/ActaHortic.2009.815.37
Ježić M, Krstin L, Rigling D, Ćurkovic-Perica M (2012) High diversity in populations of the introduced plant pathogen, Cryphonectria parasitica, due to encounters between genetically divergent genotypes. Mol Ecol 21:87–99. https://doi.org/10.1111/j.1365-294X.2011.05369.x
Ježić M, Krstin L, Poljak I, Liber Z, Idžojtić M, Jelić M et al (2014) Castanea sativa: genotype-dependent recovery from chestnut blight. Tree Genet Genomes 10:101–110. https://doi.org/10.1007/s11295-013-0667-z
Krstin L, Katanić Z, Ježić M, Poljak I, Nuskern L, Matković I, Idžojtić M, Ćurković-Perica M (2017) Biological control of chestnut blight in Croatia: an interaction between host sweet chestnut, its pathogen Cryphonectria parasitica and the biocontrol agent Cryphonectria hypovirus 1. Pest Manag Sci 73:582–589. https://doi.org/10.1002/ps.4335
Allemann C, Hoegger P, Heiniger U, Rigling D (1999) Genetic variation of Cryphonectria hypoviruses (CHV1) in Europe, assessed using restriction fragment length polymorphism (RFLP) markers. Mol Ecol 8:843–854. https://doi.org/10.1046/j.1365-294X.1999.00639.x
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: Molecular evolutionary genetics analysis Version 6.0. Mol Biol Evol 30:2725–2729. https://doi.org/10.1093/molbev/mst197
Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl. Acids. Symp. Ser. 41:95–98
Rozas J, Ferrer-Mata A, Sánchez-DelBarrio JC, Guirao-Rico S, Librado P, Ramos-Onsins SE, Sánchez-Gracia A (2017) DnaSP 6: DNA Sequence polymorphism analysis of large datasets. Mol Biol Evol. 34:3299–3302. https://doi.org/10.1093/molbev/msx248
Felsenstein J (1985) Phylogenies and the Comparative Method. Am Nat 125:1–15
Bandelt HJ, Forster P, Röhl A (1999) Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol 16:37–48. https://doi.org/10.1093/oxfordjournals.molbev.a026036
Martin D, Murrell B, Golden M, Khoosal A, Muhire B (2015) RDP4: detection and analysis of recombination patterns in virus genomes. Virus Evol 1. https://doi.org/10.1093/ve/vev003
Rice P, Longden I, Bleasby A (2000) EMBOSS: The European Molecular Biology Open Software Suite. Trends Genet 16:276–277. https://doi.org/10.1016/S0168-9525(00)02024-2
Goujon M, McWilliam H, Li W, Valentin F, Squizzato S, PaernJ, et al (2010) A new bioinformatics analysis tools framework at EMBL-EBI. Nucleic Acids Res. https://doi.org/10.1093/nar/gkq313
Waterhouse AM, Procter JB, Martin DMA, Clamp M, Barton GJ (2009) Jalview Version 2—a multiple sequence alignment editor and analysis workbench. Bioinformatics 25:1189–1191. https://doi.org/10.1093/bioinformatics/btp033
Livingstone CD, Barton GJ (1993) Protein sequence alignments: a strategy for the hierarchical analysis of residue conservation. Bioinformatics 9:745–756. https://doi.org/10.1093/bioinformatics/9.6.745
Hillman BI, Shapira R, Nuss DL (1990) Hypovirulence-associated suppression of host functions in Cryphonectria parasitica can be partially relieved by high light intensity. Phytopathology 80:950–956. https://doi.org/10.1094/Phyto-80-950
Zhu MF, Dong J and Cao DS (2016) rDNAse: generating various numerical representation schemes of DNA sequences. R package version 1.1–1. https://CRAN.R-project.org/package=rDNAse.
Li H (2018) BioSeqClass: classification for biological sequences. R package version 1.40.0. http://master.bioconductor.org/packages//2.10/bioc/html/BioSeqClass.html.
Kawashima S, Ogata H, Kanehisa M (1999) AAindex: amino acid index database. Nucleic Acids Res 27:368–369. https://doi.org/10.1093/nar/27.1.368
Kuhn M, contributions from Wing J, Weston S, Williams A, Keefer C, Engelhardt A, Cooper T, Mayer Z, Kenkel B, the R Core Team, Benesty M, Lescarbeau R, Ziem A, Scrucca L, Tang Y, Candan C and Hunt T (2018) caret: Classification and regression training. R package version 6.0–81. https://CRAN.R-project.org/package=caret.
Choi GH, Pawlyk DM, Nuss DL (1991) The autocatalytic protease p29 encoded by a hypovirulence-associated virus of the chestnut blight fungus resembles the potyvirus-encoded protease HC-Pro. Virology 183:747–752. https://doi.org/10.1016/0042-6822(91)91004-Z
Duffy S, Shackelton LA, Holmes EC (2008) Rates of evolutionary change in viruses: patterns and determinants. Nat Rev Genet 9:267–276. https://doi.org/10.1038/nrg2323
Halambek M (1986) Chestnut blight in Yugoslavia. EPPO Bull 16:533–535
Halambek M (1988) Istraživanje virulentnost i gljive Endothia parasitica Murr./And. uzročnika raka kore pitomog kestena (Castaneasativa Mill.). Dissertation. Faculty of Forestry, University of Zagreb, Croatia
Liu YC, Dynek JN, Hillman BI, Milgroom MG (2007) Diversity of viruses in Cryphonectria parasitica and C. nitschkei in Japan and China, and partial characterization of a new chrysovirus species. Mycol Res 111:433–442. https://doi.org/10.1016/j.mycres.2006.12.006
Pérez-Losada M, Arenas M, Galán JC, Palero F, González-Candelas F (2015) Recombination in viruses: mechanisms, methods of study, and evolutionary consequences. Infect Genet Evol 30:296–307. https://doi.org/10.1016/j.meegid.2014.12.022
Domingo E, Holland JJ (1997) RNA virus mutations and fitness for survival. Annu Rev Microbiol 51:151–178. https://doi.org/10.1146/annurev.micro.51.1.151
Lima ATM, Silva JCF, Silva FN, Castillo-Urquiza GP, Silva FF, Seah YM et al (2017) The diversification of begomovirus populations is predominantly driven by mutational dynamics. Virus Evol. https://doi.org/10.1093/ve/vex005
Khalifa ME, Pearson MN (2014) Characterisation of a novel hypovirus from Sclerotinia sclerotiorum potentially representing a new genus within the Hypoviridae. Virology 464–465:441–449 https://doi.org/10.1016/j.virol.2014.07.005
Du Y, Lin Y, Zhou X, Wang K, Fang S, Deng Q (2017) Full-length sequence and genome analysis of CHV1-CN280, a North China isolate of Cryphonectri ahypovirus 1. Arch Virol 162:1811–1818. https://doi.org/10.1007/s00705-017-3296-2
Nuskern L, Tkalec M, Ježić M, Katanić Z, Krstin L, Ćurković-Perica M (2017) Cryphonectria hypovirus 1-induced changes of stress enzyme activity in transfected phytopathogenic fungus Cryphonectria parasitica. Microb Ecol 74:302–311. https://doi.org/10.1007/s00248-017-0945-7
Shabalina SA, Spiridonov NA, Kashina A (2013) Sounds of silence: synonymous nucleotides as a key to biological regulation and complexity. Nucleic Acids Res 41:2073–2094. https://doi.org/10.1093/nar/gks1205
Martínez MA, Jordan-Paiz A, Franco S, Nevot M (2016) Synonymous virus genome recoding as a tool to impact viral fitness. Curr Trends Microbiol 24:134–147. https://doi.org/10.1016/j.tim.2015.11.002
Acknowledgments
This research was supported by the Swiss National Science Foundation and by Josip Juraj Strossmayer University of Osijek (project IZIP-2016-54). We thank Dr. Daniel Rigling in whose laboratory at the WSL (Switzerland) part of the experimental work was conducted.
Author information
Authors and Affiliations
Corresponding author
Additional information
Ljiljana Krstin and Zorana Katanić should be considered joint first author.
Rights and permissions
About this article
Cite this article
Krstin, L., Katanić, Z., Repar, J. et al. Genetic Diversity of Cryphonectria hypovirus 1, a Biocontrol Agent of Chestnut Blight, in Croatia and Slovenia. Microb Ecol 79, 148–163 (2020). https://doi.org/10.1007/s00248-019-01377-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00248-019-01377-9