Abstract
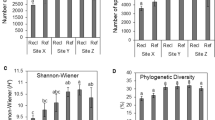
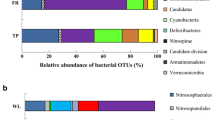
Soil microorganisms are sensitive to environmental perturbations such that changes in microbial community structure and function can provide early signs of anthropogenic disturbances and even predict restoration success. We evaluated the bacterial functional diversity of un-mined and three chronosequence sites at various stages of rehabilitation (0, 10, and 20 years old) located in the Mocho Mountains of Jamaica. Samples were collected during the dry and wet seasons and analyzed for metal concentrations, microbial biomass carbon, bacterial numbers, and functional responses of soil microbiota using community-level physiological profile (CLPP) assays. Regardless of the season, un-mined soils consisted of higher microbial biomass and numbers than any of the rehabilitated sites. Additionally, the number and rate of substrates utilized and substrate evenness (the distribution of color development between the substrates) were significantly greater in the un-mined soils with carbohydrates being preferentially utilized than amino acids, polymers, carboxylic acids, and esters. To some extent, functional responses varied with the seasons but the least physiological activity was shown by the site rehabilitated in 1987 indicating long-term perturbation to this ecosystem. Small subunit ribosomal DNA (SSUrDNA)-denaturing gradient-gel electrophoresis analyses on the microbiota collected from the most preferred CLPP substrates followed by taxonomic analyses showed Proteobacteria, specifically the gamma-proteobacteria, as the most functionally active phyla, indicating a propensity of this phyla to out-compete other groups under the prevailing conditions. Additionally, multivariate statistical analyses, Shannon's diversity, and evenness indices, principal component analysis, biplot and un-weighted-pair-group method with arithmetic averages dendrograms further confirmed that un-mined sites were distinctly different from the rehabilitated soils.








Similar content being viewed by others
References
Anderson JD, Ingram LJ, Stahl PD (2008) Influence of reclamation management practices on microbial biomass carbon soil organic carbon accumulation in semiarid mined lands of Wyoming. Appl Soil Ecol 40:387–397
Anderson JD, Stahl PD, Mummey DL (2002) Indicators of mine soil recovery. Proceedings of the Western Soil Science Society 7th Annual Meeting Colorado State University, Fort Collins
Chauhan A, Cherrier J, Williams HN (2009) Impact of sideways and bottom up control factors on bacterial community succession over a tidal cycle. Proc Natl Acad Sci USA 106(11):4301–4306
Chauhan A, Fortenberry G, Lewis D, Williams HN (2009) Increased diversity of predacious Bdellovibrio-like organisms as a function of eutrophication in Kumaon Lakes of India. Curr Microbiol 59(1):1–8
Chauhan A, Ogram AV, Reddy KR (2004) Syntrophic-methanogenic associations along a nutrient gradient in the Florida Everglades. Appl Environ Microbiol 70:3475–3484
Chauhan A, Ogram AV, Reddy KR (2006) Syntrophic archaeal associations differ with nutrient impact in a freshwater marsh. J Appl Microbiol 100:73–84
Classen AT, Boyle SI, Haskins KE, Overby ST, Hart ST (2003) Community-level physiological profiles of bacteria and fungi: plate type and incubation temperature influences on contrasting soils. FEMS Microbiol Ecol 44:319–328
Crecchio C, Gelsomino A, Ambrosoli R, Minati JL, Ruggiero P (2004) Functional and molecular responses of soil microbial communities under differing soil management practices. Soil Biol and Biochem 36:1873–1883
Doran JW, Parkin TB (1994) Defining and assessing soil quality. In: Doran JW, Coleman DC, Bezdicek DF, Stewart BA (eds) Defining soil quality for a sustainable environment. SSSA, Inc., Madison, Wisconsin, USA
Doran JW, Sarrantonio M, Liebig MA (1996) Soil health and sustainability. Adv Agron 56:1–54
Durall DM, Parsons WFJ, Parkinson D (1984) Decomposition of timothy litter (Phleum pratense) litter on a reclaimed surface coal mine in Alberta. Canada Can J Bot 36:1586–1597
Dyck WJ, Cole DW (1994) Strategies for determining consequences of harvesting and associated practices on long-term productivity. In: Dyck WJ, Cole DW (eds) Impacts of forest harvesting on long-term site productivity. Chapman & Hall, New York, pp 11–40
Garland J (1997) Analysis and interpretation of community-level physiological profiles in microbial ecology. FEMS Microbiol Ecol 24:289–300
Garland JL, Mills AL (1991) Classification and characterization of heterotrophic microbial communities on the basis of patterns of community-level sole-carbon-source utilization. Appl and Environ Microbiol 57:2351–2359
Gomez E, Ferreras L, Toresani S (2006) Soil bacterial functional diversity as influenced by organic amendment application. Biores Technoo 97:1484–1489
Gomez E, Garland J, Conti M (2004) Reproducibility in the response of soil bacterial community-level physiological profiles from a land use intensification gradient. Appl Soil Ecol 26:21–30
Graham MH, Haynes RJ (2004) Organic matter status and the size, activity and metabolic diversity of the soil microflora as indicators of the success of rehabilitation of mined sand dunes. Biol and Fert of Soils 39:429–437
Han X, Wang R, Liu J, Wang M, Zhou J, Guo W (2007) Effects of vegetation type on soil microbial community structure and catabolic diversity assessed by polyphasic methods in North China. J of Environ Sci 19:1228–1234
Harris J (2009) Soil microbial communities and restoration ecology: facilitators or followers? Science 325(5940):573–574
Hitzl W, Rangger A, Sharma S, Insam H (1997) Separation power of the 95 substrates of the BIOLOG system determined in various soils. FEMS Microbiol Ecol 22:167–174
Insam H (1997) A new set of substrates proposed for community characterization in environmental samples. In: Insam H, Rangger A (eds) Microbial communities. Functional versus structural approaches. Springer, Berlin, pp 260–261
Jasper DA (2007) Beneficial soil microorganisms of the Jarrah forest and their recovery in bauxite Southwestern Australia. Restoration Ecology 15:S74–S84
Klee AJ (1993) A computer program for the determination of most probable number and its confidence limits. J Microbiol Method 18:91–98
Kovach WL (1999) MVSP—A multivariate statistical package for Windows, Version 3.0. Kovach Computing Services, Pentraeth, Wales, UK
Lalor BM, Cookson WR, Murphy DV (2007) Comparison of two methods that assess soil community level physiological profiles in a forest ecosystem. Soil Biol Biochem 39:454–462
Lazareno S (1994) GraphPad Prism (version 1.02): produced by GraphPad Software Inc. USA. Trends in Pharmacol Sci 15:353–354
Mummey DL, Stahl PD, Buyer JS (2002) Microbial biomarkers as an indicator of ecosystem recovery following surface mine reclamation. Appl Soil Ecol 21:251–259
Mummey DL, Stahl PD, Buyer JS (2002) Soil microbiological properties 20 years after surface mine reclamation: spatial analysis of reclaimed and undisturbed sites. Soil Biol Biochem 34:1717–1725
Muyzer G, de Waal E (1994) Determination of the genetic diversity of microbial communities using DGGE analysis of PCR-amplified 16S rDNA. Microb Mats 35:207–213
Padmanabhan P, Padmanabhan S, DeRito C et al (2003) Respiration of 13C-labeled substrates added to soil in the field and subsequent 16S rRNA gene analysis of 13C-labeled soil DNA. Appl Environ Microbiol 69:1614–1622
Scholten JJ, Andriesse W (1986) Morphology, genesis and classification of three soils over limestone, Jamaica. Geoderma 39:1–40
Smalla K, Wachtendorf U, Heuer H, Liu W, Forney L (1998) Analysis of BIOLOG GN substrate utilization patterns by microbial communities. Appl and Environ Microbiol 64:1220–1225
Smith JM, Castro H, Ogram A (2007) Structure and function of methanogens along a short-term restoration chronosequence in the Florida Everglades. Appl Environ Microbiol 73(13):4135–4141
Spain AM, Krumholz LR, Elshahed MS (2009) Abundance, composition, diversity and novelty of soil proteobacteria. ISME J 3(8):992–1000
Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: Molecular evolutionary genetics analysis (MEGA) software version 4.0. Molec Biol and Evo 24:1596–1599
Vance ED, Brookes PC, Jenkinson DS (1987) An extraction method for measuring soil microbial biomass C. Soil Biol Biochem 19:703–707
White JR, Reddy KR (2000) The effects of phosphorus loading on organic nitrogen mineralization of soils and detritus along a nutrient gradient in the northern Everglades, Florida. Soil Sci Soc of America J 64:1525–1534
Winding A, Hendriksen NB (1997) Biolog substrate utilization assay for metabolic fingerprints of soil bacteria: incubation effects. In: Insam H, Rangger A (eds) Microbial communities. Functional versus structural approaches. Springer-Verlag KG Berlin, Berlin, pp 195–205
Acknowledgements
Funding for this work was provided by Jamalco/Alcoa Inc. We are also grateful to John Gardner and Hopeton Ferguson for their continued help and the Jamalco staff for providing help during sample collection. We would also like to acknowledge Maverick and Benjamin LeBlanc for their assistance in soil characterization and Sydia Williams for GIS maps of the sample locations.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary Figure 1
Percentage composition of phylogenetic groups that were found to be metabolically active in the Mocho Mountain area of Jamaica (DOC 27 kb)
Supplementary Figure 2
Phylogenetic tree of partial 16S rRNA gene sequences from domain Bacteria, identified from the predominantly utilized substrates in the CLPP assay on the un-mined and rehabilitated soils. The phylotree was constructed by the neighbor-joining method using MEGA4 software. Halobacterium salinarum was used as an outgroup (DOC 62 kb)
Supplementary Table 1
Soil microbiota identified from the most preferred BIOLOG® substrates in the CLPP assay (DOC 98 kb)
Rights and permissions
About this article
Cite this article
Lewis, D.E., White, J.R., Wafula, D. et al. Soil Functional Diversity Analysis of a Bauxite-Mined Restoration Chronosequence. Microb Ecol 59, 710–723 (2010). https://doi.org/10.1007/s00248-009-9621-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00248-009-9621-x