Abstract
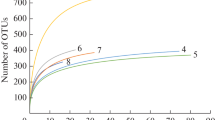
Assimilatory nitrate reductase gene fragments were isolated from epiphytes and plankton associated with seagrass blades collected from Tampa Bay, Florida, USA. Nitrate reductase genes from diatoms (NR) and heterotrophic bacteria (nasA) were amplified by polymerase chain reaction (PCR) using two sets of degenerate primers. A total of 129 NR and 75 nasA clones from four clone libraries, two from each of epiphytic and planktonic components, were sequenced and aligned. In addition, genomic DNA sequences for the NR fragment were obtained from Skeletonema costatum and Thalassiosira weissflogii diatom cultures. Rarefaction analysis with an operational taxonomic unit cut-off of 6% indicated that diversity of the NR and nasA clone libraries were similar, and that sequencing of the clone libraries was not yet saturated. Phylogenetic analysis indicated that 121 of the 129 NR clones sequenced were similar to diatom sequences. Of the eight non-diatom sequences, four were most closely related to the sequence of Chlorella vulgaris. Introns were found in 8% of the Tampa Bay NR sequences; introns were also observed in S. costatum, but not T. weissflogii. Introns from within the same clone library exhibited close similarity in nucleotide sequence, position and length; the corresponding exon sequences were unique. Introns from within the same component were similar in position and length, but not in nucleotide sequence. These findings raise questions about the function of introns, and mechanisms or time evolution of intron formation. A large cluster of 14 of the 75 nasA sequences was similar to sequences from Vibrio species; other sequences were closely related to sequences from Alteromonas, alpha-proteobacteria and Marinomonas-like species. Biogeographically consistent patterns were observed for the nasA Tampa Bay sequences compared with sequences from other locations: for example, Tampa Bay sequences were similar to those from the South Atlantic Bight, but not the Barents Sea. The Tampa Bay NR clone libraries contained sequences that exhibited phylogenetic similarity with sequences from coastal New Jersey and Monterey Bay, USA. For both NR and nasA, the sequences formed phylogenetic clusters containing nitrate reductase gene fragments that were common to both plankton and epiphyte components, and sequences that were unique to just one component. The implication that some organisms may be differentially represented in epiphytic versus planktonic components of the community suggests that local environmental conditions may have ramifications for regulation of nitrate assimilation processes, community composition, and ecosystem function.



Similar content being viewed by others
References
Allen, AA, Booth, MG, Frischer, ME, Verity, PG, Zehr, JP, Zani, S (2001) Diversity and detection of nitrate assimilation genes in marine bacteria. Appl Environ Microbiol 67: 5343–5348
Allen, AE, Booth, MG, Verity, PG, Frischer, ME (2005) Influence of nitrate availability on the distribution and abundance of heterotrophic bacterial nitrate assimilation genes in the Barents Sea during summer. Aquat Microb Ecol 39: 247–255
Allen, AE, Song, B, Ward, BB (2005) Characterisation of diatom (Bacillariophyceae) nitrate reductase genes and their detection in marine phytoplankton communities. J Phycol 41: 95–104
Bintz, JC, Nixon, SW, Buckley, BA, Granger, SL (2003) Impacts of temperature and nutrients on coastal lagoon plant communities. Estuaries 26: 765–776
Breathnach, R, Benoist, C, O’Hare, K, Gannon, F, Chambon, P (1978) Ovalbumin gene: evidence for a leader sequence in mRNA and DNA sequences at the exon–intron boundaries. Proc Natl Acad Sci USA 75: 4853–4857
Cebrian, J, Duarte, CM (2001) Detrital stocks and dynamics of the seagrass Posidonia oceanica (L.) Delile in the Spanish Mediterranean. Aquat Bot 70: 295–309
Cornelisen, C, Thomas, FIM (2002) Ammonium uptake by seagrass epiphytes: isolation of the effects of water velocity using an isotope label. Limnol Oceanogr 47: 1223–1229
Cornelisen, CD (2003) Nutrient Uptake By Seagrass Communities And Associated Organisms: Impact Of Hydrodynamic Regime Quantified Through Field Measurements And Use Of An Isotope Label, Ph.D. Dissertation, University of South Florida
Cornelisen, CD, Thomas, FIM (2004) Ammonium and nitrate uptake by leaves of the seagrass Thalassia testudinum: effects of hydrodynamic regime and epiphyte cover on uptake rates. J Mar Syst 49: 177–194
den Hartog, C (1977) Structure, function, and classification in seagrass communities. In: McRoy, CP, Helfferich, C (Eds.) Seagrass Ecosystems: A Scientific Perspective, Marine Science, vol. 4, chap. 3, Marcel Dekker, USA
Denisov, DA, Denisova, GF (2003) Hierarchical structure of donor splice site sequence multitude. Gene 320: 89–96
Duarte, CM (1995) Submerged aquatic vegetation in relation to different nutrient regimes. Ophelia 41: 87–112
Duarte, CM, Chiscano, CL (1999) Seagrass biomass and production: a reassessment. Aquat Bot 59: 21–36
Dudley, BJ, Gahnstrom, AME, Walker, DI (2001) The role of benthic vegetation as a sink for elevated inputs of ammonium and nitrate in a mesotrophic estuary. Mar Ecol Prog Ser 219: 99–107
Fonseca, MS, Fisher, JS (1986) A comparison of canopy friction and sediment movement between four species of seagrass with reference to their ecology and restoration. Mar Ecol Prog Ser 29: 15–22
Harlin, MM (1980) Seagrass epiphytes. In: Phillips, RC, McRoy, CP (Eds.) Handbook of Seagrass Biology: An Ecosystem Perspective, chap. 8, Garland STPM Press, USA
Hemminga, M, Duarte, CM (2000) Seagrass Ecology. Cambridge University Press, Cambridge, UK
Hemminga, MA, Harrison, PG, van Lent, F (1991) The balance of nutrient losses and gains in seagrass meadows. Mar Ecol Prog Ser 71: 85–96
Joint, I, Henriksen, P, Fonnes, GA, Bourne, D, Thingstad, TF, Riemann, B (2002) Competition for inorganic nutrients between phytoplankton and bacterioplankton in nutrient manipulated mesocosms. Aquat Microb Ecol 29: 145–159
Karp-Boss, L, Boss, E, Jumars, P (1996) Nutrient fluxes to planktonic osmotrophs in the presence of fluid motion. In: Ansell, AD, Gibson, RN, Barnes, MB (Eds.) Oceanography and Marine Biology: An Annual Review, UCL Press, pp 71–107
Kenworthy, WJ, Zieman, JC, Thayer, GW (1982) Evidence for the influence of seagrasses on the benthic nitrogen cycle in a coastal plain estuary near Beaufort, North Carolina (USA). Oecologia 54: 152–158
Lapointe, BE, Barile, PJ, Matzie, WR (2004) Anthropogenic nutrient enrichment of seagrass and coral reef communities in the Lower Florida Keys: discrimination of local versus regional nitrogen sources. J Exp Mar Biol Ecol 308: 23–58
Lee, KS, Dunton, KH (1999) Inorganic nitrogen acquisition in the seagrass Thalassia testudinum: development of a whole plant budget. Limnol Oceanogr 44: 1204–1215
McRoy, CP, Barsdate, RJ (1970) Phosphate absorption in Eelgrass. Limnol Oceanogr 15: 6–13
Moncreiff, CA, Sullivan, MJ, Daehnick, AE (1992) Primary production dynamics in seagrass beds of Mississippi Sound: the contributions of seagrass, epiphytic algae, sand micro?ora, and phytoplankton. Mar Ecol Prog Ser 87: 161–171
Mount, SM (1982) A catalogue of splice junction sequences. Nucleic Acids Res 10: 459–472
Papadimitriou, S, Kennedy, H, Kennedy, DP (2005) Sources of organic matter in seagrass colonized sediments: a stable isotope study of the silt and clay fraction from Posidonia oceanica meadows in the western Mediterranean. Org Geochem 36: 949–961
Senapathy, P, Shapiro, MB, Harris, NL (1990) Splice junctions, branch point sites, and exons: sequence statistics, identification and genome project. Methods Enzymol 183: 252–278
Short, FT, Short, CA (1984) The seagrass filter: purification of estuarine and coastal waters. In: Kennedy, VC (Ed.) The Estuary as a Filter, Academic Press, Orlando, FL
Tindall, KR, Kunkel, TT (1988) Fidelity of DNA synthesis by the Thermus aquaticus DNA polymerase. Biochemistry 27: 6008–6013
Valiela, I, McClelland, J, Hauxwell, J (1997) Macroalgal blooms in shallow estuaries: controls and ecophysiological and ecosystem consequences. Limnol Oceanogr 42: 1105–1118 (Part 2)
Vissini, S, Sara, G, Michener, R, Mazzola, A (2002) The role and contribution of the seagrass Posidonia oceanica (L.) Delile organic matter for secondary consumers revealed by carbon and nitrogen isotope analysis. Acta Oecol 23: 277–285
Walker, DI, McComb, AJ (1992) Seagrass degradation in Australian coastal waters. Mar Pollut Bull 25: 191–195
Young, EB, Lavery, PS, van Elven, B, Dring, MJ, Berges, JA (2005) Nitrate reductase activity in macroalgae and its vertical distribution in macroalgal epiphytes of seagrasses. Mar Ecol Prog Ser 288: 103–114
Acknowledgments
The authors thank Sean Kinane, Mark Driscoll, and Alison Meyers for field assistance; Bongkeun Song for providing Thalassiosira weissflogii DNA; and Andrew Allen for assistance with NR and nasA database sequences. This work was supported by NSF grant OCE9981482 to BBW, NSF grants and OCE-0424978 and OCE-0337052 to FIMT, and USGS cooperative agreement 04ERAAG0050 to FIMT.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Adhitya, A., Thomas, F.I.M. & Ward, B.B. Diversity of Assimilatory Nitrate Reductase Genes From Plankton and Epiphytes Associated with a Seagrass Bed. Microb Ecol 54, 587–597 (2007). https://doi.org/10.1007/s00248-006-9175-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00248-006-9175-0