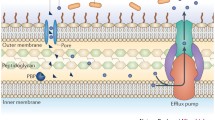
Abstract
In recent years, we have seen antimicrobial resistance rapidly emerge at a global scale and spread from one country to the other faster than previously thought. Superbugs and multidrug-resistant bacteria are endemic in many parts of the world. There is no question that the widespread use, overuse, and misuse of antimicrobials during the last 80 years have been associated with the explosion of antimicrobial resistance. On the other hand, the molecular pathways behind the emergence of antimicrobial resistance in bacteria were present since ancient times. Some of these mechanisms are the ancestors of current resistance determinants. Evidently, there are plenty of putative resistance genes in the environment, however, we cannot yet predict which ones would be able to be expressed as phenotypes in pathogenic bacteria and cause clinical disease. In addition, in the presence of inhibitory and sub-inhibitory concentrations of antibiotics in natural habitats, one could assume that novel resistance mechanisms will arise against antimicrobial compounds. This review presents an overview of antimicrobial resistance mechanisms, and describes how these have evolved and how they continue to emerge. As antimicrobial strategies able to bypass the development of resistance are urgently needed, a better understanding of the critical factors that contribute to the persistence and spread of antimicrobial resistance may yield innovative perspectives on the design of such new therapeutic targets.
Similar content being viewed by others
References
Abraham EP, Chain E (1940) An enzyme from bacteria able to destroy penicillin [1]. Nature. https://doi.org/10.1038/146837a0
Aldred KJ, Kerns RJ, Osheroff N (2014) Mechanism of quinolone action and resistance. Biochemistry. https://doi.org/10.1021/bi5000564
Allen HK, Moe LA, Rodbumrer J, Gaarder A, Handelsman J (2009) Functional metagenomics reveals diverse Β-lactamases in a remote Alaskan soil. ISME J. https://doi.org/10.1038/ismej.2008.86
Andersson DI, Hughes D (2011) Peristence of antibiotic resistance in bacterial populations. FEMS Microbiol Rev 35:901–911
Andersson DI, Cars O, Runehagen A, Sjolund-Karlsson M, Cars H, Sundqvist M et al (2009) Little evidence for reversibility of trimethoprim resistance after a drastic reduction in trimethoprim use. J Antimicrob Chemother 65(2):350–360. https://doi.org/10.1093/jac/dkp387
Aroniadis OC, Brandt LJ (2013) Fecal microbiota transplantation: past, present and future. Curr Opin Gastroenterol 29(1):79–84. https://doi.org/10.1097/MOG.0b013e32835a4b3e
Austin M, Mellow M, Tierney WM (2014) Fecal microbiota transplantation in the treatment of clostridium difficile infections. Am J Med. https://doi.org/10.1016/j.amjmed.2014.02.017
Bahl MI, Hansen LH, Sørensen SJ (2009) Persistence mechanisms of conjugative plasmids. Methods Mol Biol 532:73–102. https://doi.org/10.1007/978-1-60327-853-9_5
Baker SJ, Payne DJ, Rappuoli R, De Gregorio E (2018) Technologies to address antimicrobial resistance. Proc Natl Acad Sci USA. https://doi.org/10.1073/pnas.1717160115
Ball PR, Shales SW, Chopra I (1980) Plasmid-mediated tetracycline resistance in escherichia coli involves increased efflux of the antibiotic. Biochem Biophys Res Commun 93(1):74–81. https://doi.org/10.1016/S0006-291X(80)80247-6
Baltz RH (2006) Marcel faber roundtable: is our antibiotic pipeline unproductive because of starvation, constipation or lack of inspiration? J Ind Microbiol Biotechnol. https://doi.org/10.1007/s10295-005-0077-9
Barbe V, Vallenet D, Fonknechten N, Kreimeyer A, Oztas S, Labarre L et al (2004) Unique features revealed by the genome sequence of Acinetobacter sp. ADP1, a versatile and naturally transformation competent bacterium. Nucleic Acids Res 1:1. https://doi.org/10.1093/nar/gkh910
Bartoloni A, Pallecchi L, Rodríguez H, Fernandez C, Mantella A, Bartalesi F et al (2009) Antibiotic resistance in a very remote Amazonas community. Int J Antimicrob Agents. https://doi.org/10.1016/j.ijantimicag.2008.07.029
Bayer AS, Schneider T, Sahl HG (2013) Mechanisms of daptomycin resistance in Staphylococcus aureus: role of the cell membrane and cell wall. Ann N Y Acad Sci 1277(1):139–158. https://doi.org/10.1111/j.1749-6632.2012.06819.x
Benveniste R, Davies J (1973) Aminoglycoside antibiotic-inactivating enzymes in actinomycetes similar to those present in clinical isolates of antibiotic-resistant bacteria. Proc Natl Acad Sci USA 70:2276–2280
Bhullar K, Waglechner N, Pawlowski A, Koteva K, Banks ED, Johnston MD et al (2012) Antibiotic resistance is prevalent in an isolated cave microbiome. PLoS ONE. https://doi.org/10.1371/journal.pone.0034953
Biliński J, Grzesiowski P, Muszyński J, Wróblewska M, Mądry K, Robak K et al (2016) Fecal microbiota transplantation inhibits multidrug-resistant gut pathogens: preliminary report performed in an immunocompromised host. Arch Immunol Ther Exp. https://doi.org/10.1007/s00005-016-0387-9
Borrell S, Teo Y, Giardina F, Streicher EM, Klopper M, Feldmann J et al (2013) Epistasis between antibiotic resistance mutations drives the evolution of extensively drug-resistant tuberculosis. Evol Med Public Health 2013(1):65–74. https://doi.org/10.1093/emph/eot003
Boucher Y, Labbate M, Koenig JE, Stokes HW (2007) Integrons: mobilizable platforms that promote genetic diversity in bacteria. Trends Microbiol. https://doi.org/10.1016/j.tim.2007.05.004
Brockhurst MA, Harrison F, Veening J-W, Harrison E, Blackwell G, Iqbal Z, Maclean C (2019) Assessing evolutionary risks of resistance for new antimicrobial therapies. Nat Ecol Evol. https://doi.org/10.1038/s41559-019-0854-x
Cantón R (2009) Antibiotic resistance genes from the environment: a perspective through newly identified antibiotic resistance mechanisms in the clinical setting. Clin Microbiol Infect. https://doi.org/10.1111/j.1469-0691.2008.02679.x
Carroll AC, Wong A (2018) Plasmid persistence: costs, benefits and the plasmid paradox. Can J Microbiol 64:293–304. https://doi.org/10.1139/cjm-2017-0609
CDC (2013) Centers for disease control and prevention (CDC). Antibiotic resistance threats in the United States, 2013. https://www.cdc.gov/drugresistance/pdf/ar-threats-2013-508.pdf. Current. https://doi.org/CS239559-B
Chambers HF (1999) Penicillin-binding protein-mediated resistance in Pneumococci and Staphylococci. J Infect Dis. https://doi.org/10.1086/513854
Connell SR, Tracz DM, Nierhaus KH, Taylor DE (2003) Ribosomal protection proteins and their mechanism of Tetracycline resistance. Antimicrob Agents Chemother. https://doi.org/10.1128/AAC.47.12.3675-3681.2003
Costelloe C, Metcalfe C, Lovering A, Mant D, Hay AD (2010) Effect of antibiotic prescribing in primary care on antimicrobial resistance in individual patients: systematic review and meta-analysis. BMJ. https://doi.org/10.1136/bmj.c2096
Courvalin P (2006) Vancomycin resistance in gram-positive cocci. Clin Infect Dis. https://doi.org/10.1086/491711
D’Costa VM, McGrann KM, Hughes DW, Wright GD (2006) Sampling the antibiotic resistome. Science 311:374–377
Dahlberg C, Chao L (2003) Amelioration of the cost of conjugative plasmid carriage in Eschericha coli K12. Genetics 165(4 PG-1641–9):1641–1649
Dalmolin TV, De Lima-Morales D, Barth AL (2018) Plasmid-mediated colistin resistance: what do we know? J Infectiol Mini Rev 1:16–22
Dantas G, Sommer MOA, Oluwasegun RD, Church GM (2008) Bacteria subsisting on antibiotics. Science. https://doi.org/10.1126/science.1155157
Datta N, Hughes VM (1983) Plasmids of the same Inc groups in enterobacteria before and after the medical use of antibiotics. Nature. https://doi.org/10.1038/306616a0
Davies J, Davies D (2010) Origins and evolution of antibiotic resistance O. Microbiol Mol Biol Rev. https://doi.org/10.1128/MMBR.00016-10
Davies J, Spiegelman GB, Yim G (2006) The world of subinhibitory antibiotic concentrations. Curr Opin Microbiol. https://doi.org/10.1016/j.mib.2006.08.006
Dcosta VM, King CE, Kalan L, Morar M, Sung WWL, Schwarz C et al (2011) Antibiotic resistance is ancient. Nature. https://doi.org/10.1038/nature10388
Di Luca MC, Sørum V, Starikova I, Kloos J, Hülter N, Naseer U et al (2017) Low biological cost of carbapenemase-encoding plasmids following transfer from Klebsiella pneumoniae to Escherichia coli. J Antimicrob Chemother 72(1):85–89. https://doi.org/10.1093/jac/dkw350
Driffield K, Miller K, Bostock JM, O’neill AJ, Chopra I (2008) Increased mutability of Pseudomonas aeruginosa in biofilms. J Antimicrob Chemother. https://doi.org/10.1093/jac/dkn044
Eliopoulos GM, Huovinen P (2001) Resistance to trimethoprim-sulfamethoxazole. Clin Infect Dis 32(11):1608–1614. https://doi.org/10.1086/320532
Enne VI, Livermore DM, Stephens P, Hall LMC (2001) Persistence of sulphonamide resistance in Escherichia coli in the UK despite national prescribing restriction. Lancet 357(9265):1325–1328. https://doi.org/10.1016/S0140-6736(00)04519-0
Enne VI, Bennett PM, Livermore DM, Hall LMC (2004) Enhancement of host fitness by the sul2-coding plasmid p9123 in the absence of selective pressure. J Antimicrob Chemother 53(6):958–963. https://doi.org/10.1093/jac/dkh217
Enne VI, Delsol AA, Davis GR, Hayward SL, Roe JM, Bennett PM (2005) Assessment of the fitness impacts on Escherichia of acquisition of antibiotic resistance genes encoded by different types of genetic element. J Antimicrob Chemother 56(3):544–551. https://doi.org/10.1093/jac/dki255
European Centre for Disease Prevention and Control (2018) Surveillance of antimicrobial resistance in Europe Annual report of the European Antimicrobial Resistance Surveillance Network (EARS-Net) 2017. ECDC: Surveill Rep. https://doi.org/10.2900/230516
Fernández L, Hancock REW (2012) Adaptive and mutational resistance: role of porins and efflux pumps in drug resistance. Clin Microbiol Rev 25(4):661–681. https://doi.org/10.1128/CMR.00043-12
Fernández L, Breidenstein EBM, Hancock REW (2011) Creeping baselines and adaptive resistance to antibiotics. Drug Resist Updates 14(1):1–21. https://doi.org/10.1016/j.drup.2011.01.001
Flensburg J, Sköld O (1987) Massive overproduction of dihydrofolate reductase in bacteria as a response to the use of trimethoprim. Eur J Biochem 162(3):473–476. https://doi.org/10.1111/j.1432-1033.1987.tb10664.x
Forsman M, Haggstrom B, Lindgren L, Jaurin B (2009) Molecular analysis of β-lactamases from four species of streptomyces: comparison of amino acid sequences with those of other β-clactamases. J Gen Microbiol. https://doi.org/10.1099/00221287-136-3-589
Gillings MR (2014) Integrons: past, present, and future. Microbiol Mol Biol Rev. https://doi.org/10.1128/MMBR.00056-13
Gillings MR, Paulsen IT, Tetu SG (2017) Genomics and the evolution of antibiotic resistance. Ann N Y Acad Sci. https://doi.org/10.1111/nyas.13268
Gniadkowski M (2008) Evolution of extended-spectrum β-lactamases by mutation. Clin Microbiol Infect. https://doi.org/10.1111/j.1469-0691.2007.01854.x
Goldstein BP (2014) Resistance to rifampicin: a review. J Antibiot. https://doi.org/10.1038/ja.2014.107
Harbarth S, Balkhy HH, Goossens H, Jarlier V, Kluytmans J, Laxminarayan R et al (2015) Antimicrobial resistance: one world, one fight! Antimicrob Resist Infect Control. https://doi.org/10.1186/s13756-015-0091-2
Harkins CP, Pichon B, Doumith M, Parkhill J, Westh H, Tomasz A et al (2017) Methicillin-resistant Staphylococcus aureus emerged long before the introduction of methicillin into clinical practice. Genome Biol. https://doi.org/10.1186/s13059-017-1252-9
Harrison E, Dytham C, Hall JPJ, Guymer D, Spiers AJ, Paterson S, Brockhurst MA (2016) Rapid compensatory evolution promotes the survival of conjugative plasmids. Mob Genet Elem 6(3):2034–2039
Hiramatsu K, Ito T, Tsubakishita S, Sasaki T, Takeuchi F, Morimoto Y et al (2013) Genomic basis for methicillin resistance in Staphylococcus aureus. Infect Chemother. https://doi.org/10.3947/ic.2013.45.2.117
Holmes AH, Moore LSP, Sundsfjord A, Steinbakk M, Regmi S, Karkey A et al (2016) Understanding the mechanisms and drivers of antimicrobial resistance. The Lancet. https://doi.org/10.1016/S0140-6736(15)00473-0
Howden BP, Davies JK, Johnson PDR, Stinear TP, Grayson ML (2010) Reduced vancomycin susceptibility in Staphylococcus aureus, including vancomycin-intermediate and heterogeneous vancomycin-intermediate strains: resistance mechanisms, laboratory detection, and clinical implications. Clin Microbiol Rev. https://doi.org/10.1128/CMR.00042-09
Jacoby GA (2009) AmpC β-Lactamases. Clin Microbiol Rev. https://doi.org/10.1128/CMR.00036-08
Jacoby GA, Strahilevitz J, Hooper D (2014) Plasmid-mediated quinolone resistance. NIH Public Access Microbiol Spectr 2(2):1–15
Joon-Hee Lee (2019) Perspectives towards antibiotic resistance: from molecules to population. J Microbiol 57(3):181–184
Jouhten H, Mattila E, Arkkila P, Satokari R (2016) Reduction of antibiotic resistance genes in intestinal microbiota of patients with recurrent clostridium difficile infection after fecal microbiota transplantation. Clin Infect Dis 63(5):710–711
Kehrenberg C, Schwarz S, Jacobsen L, Hansen LH, Vester B (2005) A new mechanism for chloramphenicol, florfenicol and clindamycin resistance: methylation of 23S ribosomal RNA at A2503. Mol Microbiol 57(4):1064–1073. https://doi.org/10.1111/j.1365-2958.2005.04754.x
Keith P (2005) Efflux-mediated antimicrobial resistance. J Antimicrob Chemother 56:20–51
Knothe H, Shah P, Krcmery V, Antal M, Mitsuhashi S (1983) Transferable resistance to cefotaxime, cefoxitin, cefamandole and cefuroxime in clinical isolates of Klebsiella pneumoniae and Serratia marcescens. Infection 11(6):315–317. https://doi.org/10.1007/BF01641355
Lakin SM, Dean C, Noyes NR, Dettenwanger A, Ross AS, Doster E et al (2017) MEGARes: an antimicrobial resistance database for high throughput sequencing. Nucleic Acids Res. https://doi.org/10.1093/nar/gkw1009
Leclercq R (2002) Mechanisms of resistance to macrolides and lincosamides: nature of the resistance elements and their clinical implications. Clin Infect Dis 34(4):482–492. https://doi.org/10.1086/324626
Levin BR, Perrot V, Walker N (2000) Compensatory mutations, antibiotic resistance and the population genetics of adaptive evolution in bacteria. Genetics 154(3):985–997. https://doi.org/10.1534/genetics.110.124628
Levine DP (2005) Vancomycin: a history. Clin Infect Dis. https://doi.org/10.1086/491709
Levin-Reisman I, Ronin I, Gefen O, Braniss I, Shoresh N, Balaban NQ (2017) Antibiotic tolerance facilitates the evolution of resistance. Science. https://doi.org/10.1126/science.aaj2191
Levin-Reisman I, Brauner A, Ronin I, Balaban NQ (2019) Epistasis between antibiotic tolerance, persistence, and resistance mutations. Proc Natl Acad Sci USA 116(29):14734–14739. https://doi.org/10.1073/pnas.1906169116
Liebert CA, Hall RM, Summers AO (1999) Transposon Tn21, flagship of the floating genome. Microbiol Mol Biol Rev 63:507–522
Lili LN, Britton NF, Feil EJ (2007) The persistence of parasitic plasmids. Genetics 177(1):399–405. https://doi.org/10.1534/genetics.107.077420
Liu YY, Wang Y, Walsh TR, Yi LX, Zhang R, Spencer J et al (2016) Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis. https://doi.org/10.1016/S1473-3099(15)00424-7
Livermore DM, Canton R, Gniadkowski M, Nordmann P, Rossolini GM, Arlet G et al (2007) CTX-M: changing the face of ESBLs in Europe. J Antimicrob Chemother 59:165–174
Luo N, Pereira S, Sahin O, Lin J, Huang S, Michel L, Zhang Q (2005) Enhanced in vivo fitness of fluoroquinolone-resistant Campylobacter jejuni in the absence of antibiotic selection pressure. Proc Natl Acad Sci 102(3):541–546. https://doi.org/10.1073/pnas.0408966102
Mazel D (2006) Integrons: agents of bacterial evolution. Nat Rev Microbiol. https://doi.org/10.1038/nrmicro1462
McArthur AG, Waglechner N, Nizam F, Yan A, Azad MA, Baylay AJ et al (2013) The comprehensive antibiotic resistance database. Antimicrob Agents Chemother. https://doi.org/10.1128/aac.00419-13
McMurry L, Petrucci RE, Levy SB (2006) Active efflux of tetracycline encoded by four genetically different tetracycline resistance determinants in Escherichia coli. Proc Natl Acad Sci 77(7):3974–3977. https://doi.org/10.1073/pnas.77.7.3974
Miller WR, Munita JM, Arias CA (2014) Mechanisms of antibiotic resistance in enterococci. Expert Rev Anti Infect Ther. https://doi.org/10.1586/14787210.2014.956092
Moellering RC (2012) MRSA: the first half century. J Antimicrob Chemother 67(1):4–11. https://doi.org/10.1093/jac/dkr437
Morales G, Picazo JJ, Baos E, Candel FJ, Arribi A, Peláez B et al (2010) Resistance to linezolid is mediated by the cfr gene in the first report of an outbreak of linezolid-resistant Staphylococcus aureus. Clin Infect Dis 50(6):821–825. https://doi.org/10.1086/650574
Motta SS, Cluzel P, Aldana M (2015) Adaptive resistance in bacteria requires epigenetic inheritance, genetic noise, and cost of efflux pumps. PLoS ONE 10(3):1–18. https://doi.org/10.1371/journal.pone.0118464
Munita JM, Arias CA (2016) Mechanisms of antibiotic resistance. HHPS Public Access Microbiol Spectr 4(2):1–37. https://doi.org/10.1128/microbiolspec.VMBF-0016-2015.Mechanisms
Nikaido H (1989) Outer membrane barrier as a mechanism of antimicrobial resistance. Antimicrob Agents Chemother 33(11):1831–1836
Nikaido H, Pagès JM (2012) Broad-specificity efflux pumps and their role in multidrug resistance of Gram-negative bacteria. FEMS Microbiol Rev. https://doi.org/10.1111/j.1574-6976.2011.00290.x
Norman A, Hansen LH, Sørensen SJ (2009) Conjugative plasmids: vessels of the communal gene pool. Philos Trans R Soc B. https://doi.org/10.1098/rstb.2009.0037
Nowak R (1994) Hungary sees an improvement in penicillin resistance. Science 264(5157):364. https://doi.org/10.1126/science.8153619
O’Neill J (2014) Antimicrobial resistance: tackling a crisis for the health and wealth of nations. Rev Antimicrob Resis 20:1–16
Ogawara H, Kawamura N, Kudo T, Suzuki KI, Nakase T (1999) Distribution of β-lactamases in actinomycetes. Antimicrob Agents Chemother 43:3014–3017
Osei Sekyere J (2018) Genomic insights into nitrofurantoin resistance mechanisms and epidemiology in clinical Enterobacteriaceae. Future Sci OA 4(5):293. https://doi.org/10.4155/fsoa-2017-0156
Pal C, Bengtsson-Palme J, Kristiansson E, Larsson DGJ (2015) Co-occurrence of resistance genes to antibiotics, biocides and metals reveals novel insights into their co-selection potential. BMC Genom 16(1):964. https://doi.org/10.1186/s12864-015-2153-5
Pallecchi L, Lucchetti C, Bartoloni A, Bartalesi F, Mantella A, Gamboa H et al (2007) Population structure and resistance genes in antibiotic-resistant bacteria from a remote community with minimal antibiotic exposure. Antimicrob Agents Chemother 51(4):1179–1184. https://doi.org/10.1128/AAC.01101-06
Parmar A, Lakshminarayanan R, Iyer A, Mayandi V, Leng Goh ET, Lloyd DG et al (2018) Design and syntheses of highly potent teixobactin analogues against Staphylococcus aureus, methicillin-resistant Staphylococcus aureus (MRSA), and vancomycin-resistant enterococci (VRE) in vitro and in vivo. J Med Chem. https://doi.org/10.1021/acs.jmedchem.7b01634
Partridge SR, Tsafnat G, Coiera E, Iredell JR (2009) Gene cassettes and cassette arrays in mobile resistance integrons: review article. FEMS Microbiol Rev. https://doi.org/10.1111/j.1574-6976.2009.00175.x
Paterson DL, Bonomo RA (2005) Extended-spectrum β-lactamases: a clinical update. Clin Microbiol Rev. https://doi.org/10.1128/CMR.18.4.657-686.2005
Penders J, Stobberingh EE (2008) Antibiotic resistance of motile aeromonads in indoor catfish and eel farms in the southern part of The Netherlands. Int J Antimicrob Agents. https://doi.org/10.1016/j.ijantimicag.2007.10.002
Piddock LJV (2006a) Clinically relevant chromosomally encoded multidrug resistance efflux pumps in bacteria. Clin Microbiol Rev 19(2):382–402
Piddock LJV (2006b) Multidrug-resistance efflux pumps—not just for resistance. Nat Rev Microbiol. https://doi.org/10.1038/nrmicro1464
Portsmouth S, van Veenhuyzen D, Echols R, Machida M, Ferreira JCA, Ariyasu M et al (2018) Cefiderocol versus imipenem-cilastatin for the treatment of complicated urinary tract infections caused by Gram-negative uropathogens: a phase 2, randomised, double-blind, non-inferiority trial. Lancet Infect Dis. https://doi.org/10.1016/S1473-3099(18)30554-1
Queenan AM, Bush K (2007) Carbapenemases: the versatile β-lactamases. Clin Microbiol Rev. https://doi.org/10.1128/CMR.00001-07
Ramirez MS, Tolmasky ME (2010) Aminoglycoside modifying enzymes. Drug Resist Updates 13(6):151–171. https://doi.org/10.1016/j.drup.2010.08.003
Rappuoli R, Bloom DE, Black S (2017) Deploy vaccines to fight superbugs. Nature. https://doi.org/10.1038/d41586-017-08323-0
Roberts MC (2005) Update on acquired tetracycline resistance genes. FEMS Microbiol Lett. https://doi.org/10.1016/j.femsle.2005.02.034
Roemhild R, Gokhale CS, Dirksen P, Blake C, Rosenstiel P, Traulsen A et al (2018) Cellular hysteresis as a principle to maximize the efficacy of antibiotic therapy. Proc Natl Acad Sci USA. https://doi.org/10.1073/pnas.1810004115
Rossolini GM, D’Andrea MM, Mugnaioli C (2008) The spread of CTX-M-type extended-spectrum β-lactamases. Clin Microbiol Infect. https://doi.org/10.1111/j.1469-0691.2007.01867.x
Salimiyan Rizi K, Ghazvini K, Noghondar M Kouhi (2018) Adaptive antibiotic resistance: overview and perspectives. J Infect Dis Ther. https://doi.org/10.4172/2332-0877.1000363
Salyers AA, Amábile-Cuevas CF (1997) MINIREVIEW why are antibiotic resistance genes so resistant to elimination? Antimicrob Agents Chemother 41(11):2321–2325
Schlüter A, Szczepanowski R, Kurz N, Schneiker S, Krahn I, Pühler A (2007) Erythromycin resistance-conferring plasmid pRSB105, isolated from a sewage treatment plant, harbors a new macrolide resistance determinant, an integron-containing Tn402-like element, and a large region of unknown function. Appl Environ Microbiol. https://doi.org/10.1128/AEM.02159-06
Schrag SJ, Perrot V, Levin BR (1997) Adaptation to the fitness costs of antibiotic resistance in Escherichia coli. Proc R Soc B 264(1386):1287–1291. https://doi.org/10.1098/rspb.1997.0178
Schwarz S, Kehrenberg C, Doublet B, Cloeckaert A (2004) Molecular basis of bacterial resistance to chloramphenicol and florfenicol. FEMS Microbiol Rev. https://doi.org/10.1016/j.femsre.2004.04.001
Sengupta S, Chattopadhyay MK, Grossart HP (2013) The multifaceted roles of antibiotics and antibiotic resistance in nature. Front Microbiol. https://doi.org/10.3389/fmicb.2013.00047
Sievert DM, Rudrik JT, Patel JB, Wilkins MJ, McDonald LC, Hageman JC (2008) Vancomycin-resistant Staphylococcus aureus in the United States, 2002-2006. Clin Infect Dis 46(5):668–674. https://doi.org/10.1086/527392
Sommer MOA, Church GM, Dantas G (2010) The human microbiome harbors a diverse reservoir of antibiotic resistance genes. Virulence 1(4):299–303. https://doi.org/10.4161/viru.1.4.12010
Stokes HW, Hall RM (1989) A novel family of potentially mobile DNA elements encoding site-specific gene-integration functions: integrons. Mol Microbiol. https://doi.org/10.1111/j.1365-2958.1989.tb00153.x
Trindade S, Sousa A, Xavier KB, Dionisio F, Ferreira MG, Gordo I (2009) Positive epistasis drives the acquisition of multidrug resistance. PLoS Genet. https://doi.org/10.1371/journal.pgen.1000578
Van Giau V, An SSA, Hulme J (2019) Recent advances in the treatment of pathogenic infections using antibiotics and nano-drug delivery vehicles. Drug Des Dev Ther. https://doi.org/10.2147/DDDT.S190577
Vogwill T, Maclean RC (2015) The genetic basis of the fitness costs of antimicrobial resistance: a meta-analysis approach. Evol Appl 8(3):284–295. https://doi.org/10.1111/eva.12202
Ward MJ, Gibbons CL, McAdam PR, van Bunnik BAD, Girvan EK, Edwards GF et al (2014) Time-scaled evolutionary analysis of the transmission and antibiotic resistance dynamics of Staphylococcus aureus clonal complex 398. Appl Environ Microbiol. https://doi.org/10.1128/AEM.01777-14
Wein T, Hülter NF, Mizrahi I, Dagan T (2019) Emergence of plasmid stability under non-selective conditions maintains antibiotic resistance. Nature Commun. https://doi.org/10.1038/s41467-019-10600-7
Whiteway J, Koziarz P, Veall J, Sandhu N, Kumar P, Hoecher B, Lambert IB (1998) Oxygen-insensitive nitroreductases: analysis of the roles of nfsA and nfsB in development of resistance to 5-nitrofuran derivatives in Escherichia coli. J Bacteriol 180(21):5529–5539
Wistrand-Yuen E, Knopp M, Hjort K, Koskiniemi S, Berg OG, Andersson DI (2018) Evolution of high-level resistance during low-level antibiotic exposure. Nat Commun 9(1):1–12. https://doi.org/10.1038/s41467-018-04059-1
World Health Organization (2017) Central Asian and Eastern European Surveillance of Antimicrobial Resistance (CAESAR). http://www.euro.who.int/en/health-topics/disease-prevention/antimicrobial-resistance/about-amr/central-asian-and-eastern-european-surveillance-of-antimicrobial-resistance-caesar
Wright GD (2007) The antibiotic resistome: the nexus of chemical and genetic diversity. Nat Rev Microbiol. https://doi.org/10.1038/nrmicro1614
Zervos MJ, Schaberg DR (1985) Reversal of the in vitro susceptibility of enterococci to trimethoprim-sulfamethoxazole by folinic acid. Antimicrob Agents Chemother 28(3):446–448. https://doi.org/10.1128/AAC.28.3.446
Funding
None to declare.
Author information
Authors and Affiliations
Corresponding author
Additional information
Handling Editor: Konstantinos Voskarides.
Rights and permissions
About this article
Cite this article
Christaki, E., Marcou, M. & Tofarides, A. Antimicrobial Resistance in Bacteria: Mechanisms, Evolution, and Persistence. J Mol Evol 88, 26–40 (2020). https://doi.org/10.1007/s00239-019-09914-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00239-019-09914-3