Abstract
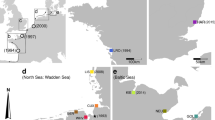
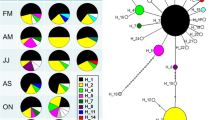
We report current genetic variation of populations of the razor shell Ensis directus (Conrad 1843) (Mollusca: Bivalvia: Pharidae) in native (North American) and introduced (European) ranges using nuclear and mitochondrial sequence-based markers. We expected less variation within the introduced range, especially considering the frequent mass mortality events observed in Europe since the species was recorded for the first time in 1978. However, we found higher variation in Europe. The possible significance of temporal fluctuations of genetic variation, limited effect of random genetic drift, and multiple introductions are discussed. Interestingly, the multiple-introduction hypothesis contrasts with the gradual colonisation of European coastal waters but is supported by trained clustering analysis and by the intensity of transatlantic shipping. Genetic and morphometric evidence strongly supports that examined individuals from a supposed E. directus population from Newfoundland (Canada) belong to a separate species. This new Ensis is formally described here and named E. terranovensis n.sp.






Similar content being viewed by others
Abbreviations
- MNCN:
-
Museo Nacional de Ciencias Naturales, Madrid, Spain
- MNHN:
-
Muséum national d’Histoire naturelle, Paris, France
- ZMUC:
-
Zoologisk Museum—Københavns Universitet, Copenhagen, Denmark
- CMNML:
-
Canadian Museum of Nature, Ottawa-Gatineau, Canada
References
April J, Mayden RL, Hanner RH, Bernatchez L (2011) Genetic calibration of species diversity among North America’s freshwater fishes. PNAS 108:10602–10607
Arias A, Anadón N (2012) First record of Mercenaria mercenaria (Bivalvia: Veneridae) and Ensis directus (Bivalvia: Pharidae) on Bay of Biscay, Iberian Peninsula. J Shellfish Res 31:57–60
Armonies W (2001) What an introduced species can tell us about the spatial extension of benthic populations. Mar Ecol Prog Ser 209:289–294
Armonies W, Reise K (1999) On the population development of the introduced razor clam Ensis americanus near the island pf Sylt (North Sea). Helgolan Mar Res 52:291–300
Arnaud-Haond S, Vonau V, Rouxel C, Bonhomme F, Prou J, Goyard E, Boudry P (2008) Genetic structure at different spatial scales in the pearl oyster (Pinctada margaritifera cumingii) in French Polynesian lagoons: beware of sampling strategy and genetic patchiness. Mar Biol 155:147–157
Audzijonyte A, Vrijenhoek RC (2010) Three nuclear genes for phylogenetic, SNP and population genetic studies of molluscs and other invertebrates. Mol Ecol Resour 10:200–204
Baker P, Austin JD, Bowen BW, Baker SM (2008) Range-wide population structure and history of the northern quahog (Merceneria merceneria) inferred from mitochondrial DNA sequence data. ICES J Mar Sci 65:155–163
Beaumont MA (2007) Conservation genetics. In: Balding DJ, Bishop M, Cannings C (eds) Handbook of statistical genetics, 3rd edn. Wiley, Chichester, pp 1021–1066
Beukema JJ, Dekker R (1995) Dynamics and growth of a recent invader into European coastal waters: the American razor clam, Ensis directus. J Mar Biol Assess UK 75:351–362
Briggs JC, Bowen BW (2012) A realignment of marine biogeographic provinces with particular reference to fish distributions. J Biogeogr 39:12–30
Bryant D, Moulton V (2004) Neighbor-net: an agglomerative method for the construction of phylogenetic networks. Mol Biol Evol 21:255–265
Cadée GC (2000) Herring gulls feeding on a recent invader in the Wadden Sea, Ensis directus. In: Harper EM, Taylor JD, Crame JA (eds), The evolutionary biology of the Bivalvia. Geol Soc Lond Special Publ 177:459–464
Cardoso JFMF, Witt JIJ, van der Veer HW (2009) Reproductive investment of the American razor clam Ensis americanus in the Dutch Wadden Sea. J Sea Res 62:295–298
Cheng L, Connor TR, Aanensen DM, Spratt BG, Corander J (2011) Bayesian semi-supervised classification of bacterial samples using MLST databases. BMC Bioinformatics 12:302. doi:10.1186/1471-2105-12-302
Clement M, Posada D, Crandall K (2000) TCS: a computer program to estimate gene genealogies. Mol Ecol 9:1657–1660
Corander J, Marttinen P, Mäntyniemi S (2006) Bayesian identification of stock mixtures from molecular marker data. Fis B-NOAA 104:550–558
Corander J, Marttinen P, Sirén J, Tang J (2008) Enhanced bayesian modelling in BAPS software for learning genetic structures of populations. BMC Bioinformatics 9:539
Cosel von R (2009) The razor shells of the eastern Atlantic, part 2. Pharidae II: the genus Ensis Schumacher, 1817 (Bivalvia, Solenoidea). Basteria 73:1–48
Dannheim J, Rumohr H (2011) The fate of an immigrant: Ensis directus in the eastern German Bight. Helgol Mar Res. doi:10.1007/s10152-011-0271-2
Dansey P (2011) Ensis directus (Conrad 1843) (Bivalvia: Solenoidea) found in Liverpool Bay (Sea area S24). J Conchol 40:679
de Queiroz K (2007) Species concepts and species delimitation. Syst Biol 56:879–886
Dekker R, Beukema JJ (2012) Long-term dynamics and productivity of a successful invader: The first three decades of the bivalve Ensis directus in the western Wadden Sea. J Sea Res http://dx.doi.org/10.1016/j.seares.2012.04.004
Dlugosch KM, Parker IM (2008) Founding events in species invasions: genetic variation, adaptive evolution, and the role of multiple introductions. Mol Ecol 17:431–449
Drummond AJ, Ashton B, Buxton S, Cheung M, Cooper A, Duran C, Field M, Heled J, Kearse M, Markowitz S, Moir R, Stones-Havas S, Sturrock S, Thierer T, Wilson A (2011) Geneious v5.4. Available from http://www.geneious.com
Excoffier L, Lischer HEL (2010) Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Res 10:564–567
Excoffier L, Smouse P, Quattro J (1992) Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics 131:479–491
Fay JC, Wu CI (2000) Hitchhiking under positive Darwinian selection. Genetics 155:1405–1413
Fitzpatrick BM, Fordyce JA, Niemiller ML, Reynolds RG (2012) What can DNA tell us about biological invasions? Biol Invasions 14:245–253
Folmer O, Black M, Hoeh W, Lutz R, Vrijenhoek R (1994) DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol Mar Biol Biotechnol 3:294–299
Freudendahl ASL, Nielsen MM, Jensen T, Jensen KT (2010) The introduced clam Ensis americanus in the Wadden Sea: field experiment on impact of bird predation and tidal level on survival and growth. Helgoland Mar Res 64:93–100
Fu YX (1997) Statistical tests of neutrality of mutations against population growth, hitchhiking, and background selection. Genetics 147:915–925
Fu YX, Li WH (1993) Statistical tests of neutrality of mutations. Genetics 133:693–709
Giegerich R, Meyer F, Schleiermacher C (1996) GeneFisher—software support for the detection of postulated genes. Proc Int Conf Intell Syst Mol Biol 4:68–77
Goudet J, Raymond M, de Meeüs T, Rousset F (1996) Testing differentiation in diploid populations. Genetics 144:1933–1940
Griffiths-Jones S (2005) RALEE—RNA ALignment editor in Emacs. Bioinformatics 21:257–259
Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O (2010) New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol 59:307–321
Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl Acids Symp Ser 41:95–98
Hare MP, Weinberg JR (2005) Phylogeography of surfclams, Spisula solidissima, in the western North Atlantic based on mitochondrial and nuclear DNA sequences. Mar Biol 146:707–716
Harpending HC (1994) Signature of ancient population growth in a low-resolution mitochondrial DNA mismatch distribution. Hum Biol 66:591–600
Heath DD, Rawson PD, Hilbish TJ (1995) PCR-based nuclear markers identify alien blue mussel (Mytilus spp.) genotypes on the west coast of Canada. Can J Fish Aquat Sci 52:2621–2627
Hedgecock D, Pudovkin AI (2011) Sweepstakes reproductive success in highly fecund marine fish and shellfish: a review and commentary. Bull Mar Sci 87:971–1002
Holland BS (2000) Genetics of marine bioinvasions. Hydrobiologia 420:63–71
Hudson R, Kreitman M, Aguadé M (1987) A test of neutral molecular evolution based on nucleotide data. Genetics 116:153–159
Huelsenbeck JP, Ronquist F (2001) MRBAYES: bayesian inference of phylogenetic trees. Bioinformatics 17:754–755
Huson DH, Bryant D (2006) Application of phylogenetic networks in evolutionary studies. Mol Biol Evol 23:254–267
Kenchington E, Duggan R, Riddell T (1998) Early life history characteristics of the razor clam (Ensis directus) and the moonsnails (Euspira spp.) with applications to fisheries and aquaculture. Can Tech Rep Fish Aquat Sci 2223:1–32
Kenchington EL, Patwary MU, Zouros E, Bird CJ (2006) Genetic differentiation in relation to marine landscape in a broadcast-spawning bivalve mollusc (Placopecten magellanicus). Mol Ecol 15:1781–1796
Kong L, Matsukuma A, Hayashi I, Takada Y, Li Q (2012) Taxonomy of Macridiscus species (Bivalvia: Veneridae) from the western Pacific: insight based on molecular evidence, with description of a new species. J Moll Stud 78:1–11
Krakau M, Thieltges DW, Reise K (2006) Native parasites adopt introduced bivalves of the North Sea. Biol Invasions 8:919–925
Krakau M, Jacobsen S, Jensen KT, Reise K (2012) The cockle Cerastoderma edule at northeast Atlantic shores: genetic signatures of glacial refugia. Mar Biol 159:221–230
Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, López R, Thompson JD, Gibson TJ, Higgins DG (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948
Librado P, Rozas J (2009) DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25:1451–1452
Luttikhuizen PC, Drent J, Baker AJ (2003) Disjunct distribution of highly diverged mitochondrial lineage clade and population subdivision in a marine bivalve with pelagic larval dispersal. Mol Ecol 12:2215–2229
Maggs CA, Castilho R, Foltz D, Henzler C, Jolly MT, Kelly J, Olsen J, Perez KE, Stam W, Väinölä R, Viard F, Wares J (2008) Evaluating signatures of glacial refugia for North Atlantic benthic marine taxa. Ecology 89:S108–S122
Maine Sea Grant (2012) Accessed at http://www.seagrant.umaine.edu/resources-for-shellfish-growers/species/razor-clam. On 01 Feb 2012
Mao Y, Gao T, Yanagimoto T, Xiao Y (2011) Molecular phylogeography of Ruditapes philippinarum in the northwestern Pacific Ocean based on COI gene. J Exp Mar Biol Ecol 407:171–181
Marine Stewardship Council (2012) Accessed at http://www.msc.org/track-a-fishery/in-assessment/north-east-atlantic/dfa-dutch-north-sea-ensis. On 01 Feb 2012
McDonald J, Kreitman M (1991) Adaptative protein evolution at adh locus in Drosophila. Nature 351:652–654
Miller MA, Pfeiffer W, Schwartz T (2010) Creating the CIPRES Science Gateway for inference of large phylogenetic trees. In: Proceedings of the gateway computing environments workshop (GCE), 14 November 2010, New Orleans, LA pp 1–8
Mühlenhardt-Siegel U, Dörjes J, von Cosel R (1983) Die amerikanische Schwertmuschel Ensis directus (Conrad) in der Deutschen Bucht: 2. Populationsdynamik. Senckenb Marit 15:93–110
Nylander JAA, Wilgenbusch JC, Warren DL, Swofford DL (2008) AWTY (are we there yet?): a system for graphical exploration of MCMC convergence in Bayesian phylogenetics. Bioinformatics 24:581–583
Palmer DW (2004) Growth of the razor clam Ensis directus, an alien species in the Wash on the east coast of England. J Mar Biol Assess UK 84:1075–1076
Raymond M, Rousset F (1995) An exact test for population differentiation. Evolution 49:1280–1283
Rogers AR, Harpending H (1992) Population growth makes waves in the distribution of pairwise genetic differences. Mol Biol Evol 9:552–569
Sargent PS, Methven DA, Hooper RG, McKenzie CH (2008) A range extension of the Atlantic silverside, Menidia menidia, to coastal waters of southwestern Newfoundland. Can Field Nat 122:338–344
Stamatakis A (2006) RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22:2688–2690
Stamatakis A, Hoover P, Rougemont J (2008) A fast bootstrapping algorithm for the RAxML web-servers. Syst Biol 57:758–771
Stephens M, Scheet P (2005) Accounting for decay of linkage disequilibrium in haplotype inference and missing-data imputation. Am J Hum Genet 76:449–462
Stephens M, Smith NJ, Donnelly P (2001) A new statistical method for haplotype reconstruction from population data. Am J Hum Genet 68:978–989
Strasser CA, Barber PH (2008) Limited genetic variation and structure in softshell clams (Mya arenaria) across their native and introduced range. Conser Genet 10:803–814
Swennen C, Leopold MF, Stock M (1985) Notes on growth and behaviour of the American razor clam Ensis directus in the Wadden Sea and the predation on it by birds. Helgolan Mar Res 39:255–261
Swofford DL (2002) PAUP*. Phylogenetic analysis using parsimony (*and other methods). Version. Sinauer Associates, Sunderland, MA, USA
Tajima F (1989) Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123:585–595
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739
Tulp I, Craeymeersch J, Leopold M, van Damme C, Fey F, Verdaat H (2010) The role of the invasive bivalve Ensis directus as food source for fish and birds in the Dutch coastal zone. Estuar Coast Shelf Sci 90:116–128
Varela MA, Martínez-Lage A, González-Tizón AM (2009) Temporal genetic variation of microsatellite markers in the razor clam Ensis arcuatus (Bivalvia: Pharidae). J Mar Biol Assess UK 89:1703–1707
Varela MA, Martínez-Lage A, González-Tizón AM (2011) Genetic heterogeneity in natural beds of the razor clam Ensis siliqua revealed by microsatellites. J Mar Biol Assess UK 92:171–177
Vierna J, Martínez-Lage A, González-Tizón AM (2010) Analysis of ITS1 and ITS2 sequences in Ensis razor shells: suitability as molecular markers at the population and species levels, and evolution of these ribosomal DNA spacers. Genome 53:23–34
von Cosel R, Dörjes J, Mühlenhardt-Siegel U (1982) Die amerikanische Schwertmuschel Ensis directus (Conrad) in der Deutschen Bucht. I. Zoogegraphis und Taxonomie im Vergleich mit den einheimischen Schwertmuschel-Arten. Senckenbergiana marit 14:147–173
Xiao J, Cordes JF, Wang H, Guo X, Reece KS (2010) Population genetics of Crassostrea ariakensis in Asia inferred from microsatellite markers. Mar Biol 157:1767–1781
Acknowledgments
We are very grateful to Rudo von Cosel for his support and his drawing of E. terranovensis holotype, and to the following colleagues who helped us in some way or another during the execution of this work: André Martel, Anja Schulze, Barbara Buge, David Palmer, Diego Fonataneto, Ferruccio Maltagliati, Horacio Naveira, Jean-Marc Gagnon, Jean-Marie Dewarumez, Jeroen Goud, Jukka Corander, Manuel Pimentel, Mark Graham, Marta Vila, Neus Marí, Nicolas Puillandre, Ole S. Tendal, Paul Dansey, Philip Sargent, Rafael Araújo, Ray J. Thompson, Robert O’Donnell, Stephen T. Tettelbach, Tim Sheehan, and Tom Schioette. We would also like to thank two anonymous reviewers for critical and helpful comments. JV has been supported by a ‘María Barbeito’ fellowship and a travel grant, both from the Consellería de Economía e Industria, Xunta de Galicia (Spain) and the European Social Fund.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by M. I. Taylor.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Vierna, J., Jensen, K.T., González-Tizón, A.M. et al. Population genetic analysis of Ensis directus unveils high genetic variation in the introduced range and reveals a new species from the NW Atlantic. Mar Biol 159, 2209–2227 (2012). https://doi.org/10.1007/s00227-012-2006-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00227-012-2006-6