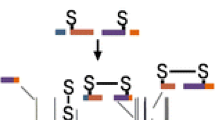
Abstract
Disulfide bonds are important structural moieties of proteins: they ensure proper folding, provide stability, and ensure proper function. With the increasing use of proteins for biotherapeutics, particularly monoclonal antibodies, which are highly disulfide bonded, it is now important to confirm the correct disulfide bond connectivity and to verify the presence, or absence, of disulfide bond variants in the protein therapeutics. These studies help to ensure safety and efficacy. Hence, disulfide bonds are among the critical quality attributes of proteins that have to be monitored closely during the development of biotherapeutics. However, disulfide bond analysis is challenging because of the complexity of the biomolecules. Mass spectrometry (MS) has been the go-to analytical tool for the characterization of such complex biomolecules, and several methods have been reported to meet the challenging task of mapping disulfide bonds in proteins. In this review, we describe the relevant, recent MS-based techniques and provide important considerations needed for efficient disulfide bond analysis in proteins. The review focuses on methods for proper sample preparation, fragmentation techniques for disulfide bond analysis, recent disulfide bond mapping methods based on the fragmentation techniques, and automated algorithms designed for rapid analysis of disulfide bonds from liquid chromatography–MS/MS data. Researchers involved in method development for protein characterization can use the information herein to facilitate development of new MS-based methods for protein disulfide bond analysis. In addition, individuals characterizing biotherapeutics, especially by disulfide bond mapping in antibodies, can use this review to choose the best strategies for disulfide bond assignment of their biologic products.

This review, describing characterization methods for disulfide bonds in proteins, focuses on three critical components: sample preparation, mass spectrometry data, and software tools








Similar content being viewed by others
References
Woycechowsky KJ, Raines RT. Native disulfide bond formation in proteins. Curr Opin Chem Biol. 2000;4(5):533–9.
Wilkinson B, Gilbert HF. Protein disulfide isomerase. Biochim Biophys Acta. 2004;1699(1-2):35–44.
Butera D, Cook KM, Chiu J, Wong JW, Hogg PJ. Control of blood proteins by functional disulfide bonds. Blood. 2014;123(13):2000–7.
Farrah T, Deutsch EW, Omenn GS, Campbell DS, Sun Z, Bletz JA, et al. A high-confidence human plasma proteome reference set with estimated concentrations in PeptideAtlas. Mol Cell Proteomics. 2011;10(9):M110.006353.
Wypych J, Li M, Guo A, Zhang Z, Martinez T, Allen MJ, et al. Structural and functional characterization of disulfide isoforms of the human IgG2 subclass. J Biol Chem. 2008;283(23):16194–205.
Alewood D, Nielsen K, Alewood PF, Craik DJ, Andrews P, Nerrie M, et al. The role of disulfide bonds in the structure and function of murine epidermal growth factor (mEGF). Growth Factors. 2005;23(2):97–110.
Zhang L, Chou CP, Moo-Young M. Disulfide bond formation and its impact on the biological activity and stability of recombinant therapeutic proteins produced by Escherichia coli expression system. Biotechnol Adv. 2011;29(6):923–9.
Perry LJ, Wetzel R. Disulfide bond engineered into T4 lysozyme: stabilization of the protein toward thermal inactivation. Science. 1984;226(4674):555–7.
Mansfeld J, Vriend G, Dijkstra BW, Veltman OR, Van den Burg B, Venema G, et al. Extreme stabilization of a thermolysin-like protease by an engineered disulfide bond. J Biol Chem. 1997;272(17):11152–6.
Kim DY, Kandalaft H, Ding W, Ryan S, van Faassen H, Hirama T, et al. Disulfide linkage engineering for improving biophysical properties of human VH domains. Protein Eng Des Sel. 2012;25(10):581–90.
Dombkowski AA, Sultana KZ, Craig DB. Protein disulfide engineering. FEBS Lett. 2014;588(2):206–12.
Seger ST, Breinholt J, Faber JH, Andersen MD, Wiberg C, Schjodt CB, et al. Probing the conformational and functional consequences of disulfide bond engineering in growth hormone by hydrogen-deuterium exchange mass spectrometry coupled to electron transfer dissociation. Anal Chem. 2015;87(12):5973–80.
Go EP, Cupo A, Ringe R, Pugach P, Moore JP, Desaire H. Native conformation and canonical disulfide bond formation are interlinked properties of HIV-1 Env glycoproteins. J Virol. 2015;90(6):2884–94.
Le QAT, Joo JC, Yoo YJ, Kim YH. Development of thermostable Candida antarctica lipase B through novel in silico design of disulfide bridge. Biotechnol Bioeng. 2012;109(4):867–76.
Compton JR, Legler PM, Clingan BV, Olson MA, Millard CB. Introduction of a disulfide bond leads to stabilization and crystallization of a ricin immunogen. Proteins. 2011;79(4):1048–60.
Liu W, Onda M, Kim C, Xiang L, Weldon JE, Lee B, et al. A recombinant immunotoxin engineered for increased stability by adding a disulfide bond has decreased immunogenicity. Protein Eng Des Sel. 2012;25(1):1–6.
Li Y, Li X, Zheng X, Tang L, Xu W, Gong M. Disulfide bond prolongs the half-life of therapeutic peptide-GLP-1. Peptides. 2011;32(7):1400–7.
Li Y, Zheng X, Tang L, Xu W, Gong M. GLP-1 analogs containing disulfide bond exhibited prolonged half-life in vivo than GLP-1. Peptides. 2011;32(6):1303–12.
Siadat OR, Lougarre A, Lamouroux L, Ladurantie C, Fournier D. The effect of engineered disulfide bonds on the stability of Drosophila Melanogaster acetylcholinesterase. BMC Biochem. 2006;7:12.
Azimi I, Wong JW, Hogg PJ. Control of mature protein function by allosteric disulfide bonds. Antioxid.Redox Signal. 2011;14(1):113–26.
Hogg PJ. Targeting allosteric disulphide bonds in cancer. Nat Rev Cancer. 2013;13(6):425–31.
Suzuki M, Yamanoi A, Machino Y, Kobayashi E, Fukuchi K, Tsukimoto M, et al. Cleavage of the interchain disulfide bonds in rituximab increases its affinity for FcγRIIIA. Biochem Biophys Res Commun. 2013;436(3):519–24.
Suzuki M, Yamanoi A, Machino Y, Ootsubo M, Izawa K, Kohroki J, et al. Effect of trastuzumab interchain disulfide bond cleavage on Fcγ receptor binding and antibody-dependent tumour cell phagocytosis. J Biochem. 2016;159(1):67–76.
Lightle S, Aykent S, Lacher N, Mitaksov V, Wells K, Zobel J, et al. Mutations within a human IgG2 antibody form distinct and homogeneous disulfide isomers but do not affect Fc gamma receptor or C1q binding. Protein Sci. 2010;19(4):753–62.
Walsh G. Biopharmaceutical benchmarks 2014. Nat Biotechnol. 2014;32(10):992–1000.
Reichert JM. Antibodies to watch in 2016. mAbs. 2016;8(2):197–204.
Michel ML, Tiollais P. Hepatitis B vaccines: protective efficacy and therapeutic potential. Pathol Biol. 2010;58(4):288–95.
Nascimento IP, Leite LCC. Recombinant vaccines and the development of new vaccine strategies. Braz J Med Biol Res. 2012;45(12):1102–11.
Amoresano A, Orru S, Siciliano RA, De Luca E, Napoleoni R, Sirna A, et al. Assignment of the complete disulphide bridge pattern in the human recombinant follitropin β-chain. Biol Chem. 2001;382(6):961–8.
Reichert JM. Marketed therapeutic antibodies compendium. mAbs. 2012;4(3):413–5.
Wadhwa M, Thorpe R. Haematopoietic growth factors and their therapeutic use. Thromb Haemost. 2008;99(5):863–73.
Calcutt NA, Jolivalt CG, Fernyhough P. Growth factors as therapeutics for diabetic neuropathy. Curr Drug Targets. 2008;9(1):47–59.
Liu HC, May K. Disulfide bond structures of IgG molecules: structural variations, chemical modifications and possible impacts to stability and biological function. mAbs. 2012;4(1):17–23.
Trexler-Schmidt M, Sargis S, Chiu J, Sze-Khoo S, Mun M, Kao YH, et al. Identification and prevention of antibody disulfide bond reduction during cell culture manufacturing. Biotechnol Bioeng. 2010;106(3):452–61.
Hutterer KM, Hong RW, Lull J, Zhao X, Wang T, Pei R, et al. Monoclonal antibody disulfide reduction during manufacturing. mAbs. 2013;5(4):608–13.
Yoshioka S, Aso Y, Izutsu K, Terao T. Aggregates formed during storage of β-galactosidase in solution and in the freeze-dried state. Pharm Res. 1993;10(5):687–91.
Chandrasekhar S, Topp EM. Thiol-disulfide exchange in peptides derived from human growth hormone during lyophilization and storage in the solid state. J Pharm Sci. 2015;104(4):1291–302.
Andya JD, Hsu CC, Shire SJ. Mechanisms of aggregate formation and carbohydrate excipient stabilizations of lyophilized humanized monoclonal antibody formulations. AAPS Pharm Sci. 2003;5(2):E10.
Vazquez-Rey M, Lang DA. Aggregates in monoclonal antibody manufacturing processes. Biotechnol Bioeng. 2011;108(7):1494–508.
Food and Drug Administration. Q6B specifications: test procedures and acceptance criteria for biotechnological/biological products. Rockville: Food and Drug Administration; 1999.
Rathore AS, Winkle H. Quality by design for biopharmaceuticals. Nat Biotechnol. 2009;27(1):26–34.
Klaus W, Broger C, Gerber P, Senn H. Determination of the disulphide bonding pattern in proteins by local and global analysis of nuclear magnetic resonance data. Application to flavoridin. J Mol Biol. 1993;232(3):897–906.
Mobli M, King GF. NMR methods for determining disulfide bond connectivities. Toxicon. 2010;56(6):849–54.
Poppe L, Hui JO, Ligutti J, Murray JK, Schnier PD. PADLOC: a powerful tool to assign disulfide bond connectivities in peptides and proteins by NMR spectroscopy. Anal Chem. 2012;84(1):262–6.
Liu D, Cowburn D. Combining biophysical methods to analyze the disulfide bond in SH2 domain of C-terminal Src kinase. Biophys Rep. 2016;2(1):33–43.
Zhang B, Cockrill SL. Methodology for determining disulfide linkage patterns of closely spaced cysteine residues. Anal Chem. 2009;81(17):7314–20.
Perham RN. A diagonal paper-electrophoretic technique for studying amino acid sequences around the cysteine and cystine residues of proteins. Biochem J. 1967;105(3):1203–7.
Milstein C. Frangion.B, Disulphide bridges of the heavy chain of human immunoglobulin G2. Biochem J. 1971;121(2):217–25.
Gorman JJ, Wallis TP, Pitt JJ. Protein disulfide bond determination by mass spectrometry. Mass Spectrom Rev. 2002;21(3):183–216.
Tsai PL, Chen SF, Huang SY. Mass spectrometry-based strategies for protein disulfide bond identification. Rev Anal Chem. 2013;32(4):257–68.
Wiesner J, Resemann A, Evans C, Suckau D, Jabs W. Advanced mass spectrometry workflows for analyzing disulfide bonds in biologics. Expert Rev Proteomics. 2015;12(2):115–23.
Wu S-L, Jiang H, Lu Q, Dai S, Hancock W, Karger BL. Mass spectrometric determination of disulfide linkages in recombinant therapeutic proteins using online LC-MS with electron-transfer dissociation. Anal Chem. 2009;81:112–22.
Clark DF, Go EP, Desaire H. Simple approach to assign disulfide connectivity using extracted ion chromatograms of electron transfer dissociation spectra. Anal Chem. 2013;85(2):1192–9.
Lakbub JC, Clark DF, Shah IS, Zhu Z, Su X, Go EP, et al. Disulfide bond characterization of endogenous IgG3 monoclonal antibodies using LC-MS: an investigation of IgG3 disulfide-mediated isoforms. Anal Methods. 2016;8(31):6046–55.
Pitt JJ, Da Silva E, Gorman JJ. Determination of the disulfide bond arrangement of Newcastle disease virus hemagglutinin neuraminidase. J Biol Chem. 2000;275(9):6469–78.
Zhang W, Marzilli LA, Rouse JC, Czupryn MJ. Complete disulfide bond assignment of a recombinant immunoglobulin G4 monoclonal antibody. Anal Biochem. 2002;311(1):1–9.
Sung WC, Chang CW, Huang SY, Wei TY, Huang YL, Lin YH, et al. Evaluation of disulfide scrambling during the enzymatic digestion of bevacizumab at various pH values using mass spectrometry. Biochim Biophys Acta. 2016;1864(9):1188–94.
Wang Y, Li H, Shameem M, Xu W. Development of a sample preparation method for monitoring correct disulfide linkages of monoclonal antibodies by liquid chromatography-mass spectrometry. Anal Biochem. 2016;495:21–8.
Sanger F. A disulphide interchange reaction. Nature. 1953;171(4362):1025–6.
Ryle AP, Sanger F. Disulphide interchange reactions. Biochem J. 1955;60(4):535–40.
Costantino HR, Schwendeman SP, Langer R, Klibanov AM. Deterioration of lyophilized pharmaceutical proteins. Biochem (Mosc). 1998;63(3):357–63.
Chandrasekhar S, Moorthy BS, Xie R, Topp EM. Thiol-disulfide exchange in human growth hormone. Pharm Res. 2016;33(6):1370–82.
Monahan FJ, German JB, Kinsella JE. Effect of pH and temperature on protein unfolding and thiol/disulfide interchange reactions during heat-induced gelation of whey proteins. J Agric Food Chem. 1995;43(1):46–52.
Kerr J, Schlosser JL, Griffin DR, Wong DY, Kasko AM. Steric effects in peptide and protein exchange with activated disulfides. Biomacromolecules. 2013;14(8):2822–9.
Wang Y, Lu Q, Wu SL, Karger BL, Hancock WS. Characterization and comparison of disulfide linkages and scrambling patterns in therapeutic monoclonal antibodies: using LC-MS with electron transfer dissociation. Anal Chem. 2011;83(8):3133–40.
Creamer LK, Bienvenue A, Nilsson H, Paulsson M, van Wanroij M, Lowe EK, et al. Heat-induced redistribution of disulfide bonds in milk proteins. 1. Bovine β-lactoglobulin. J Agric Food Chem. 2004;52(25):7660–8.
Sechi S, Chait BT. Modification of cysteine residues by alkylation. A tool in peptide mapping and protein identification. Anal Chem. 1998;70(24):5150–8.
Rogers LK, Leinweber BL, Smith CV. Detection of reversible protein thiol modifications in tissues. Anal Biochem. 2006;358(2):171–84.
Lu S, Fan S-B, Yang B, Li Y-X, Meng J-M, Wu L, et al. Mapping native disulfide bonds at the proteome scale. Nat Methods. 2015;12(4):329–31.
Switzar L, Giera M, Niessen WMA. Protein digestion: an overview of the available techniques and recent developments. J Proteome Res. 2013;12(3):1067–77.
Ni W, Lin M, Salinas P, Savickas P, Wu SL, Karger BL. Complete mapping of a cystine knot and nested disulfides by recombinant human arylsulfatase A by multi-enzyme digestion and LC-MS analysis using CID and ETD. J Am Soc Mass Spectrom. 2013;24(1):125–33.
Na S, Paek E, Choi J-S, Kim D, Lee SJ, Kwon J. Characterization of disulfide bonds by planned digestion and tandem mass spectrometry. Mol Biosyst. 2015;11(4):1156–64.
Glatter T, Ludwig C, Ahrné E, Aebersold R, Heck AJR, Schmidt A. Large-scale quantitative assessment of different in-solution protein digestion protocols reveals superior cleavage efficiency of tandem Lys-C/trypsin protocols over trypsin digestion. J Proteome Res. 2012;11(11):5145–56.
Liu F, van Breukelen B, Heck AJ. Facilitating protein disulfide mapping by a combination of pepsin digestion, electron transfer higher energy dissociation (EThcD), and a dedicated search algorithm SlinkS. Mol Cell Proteomics. 2014;13(10):2776–86.
Go EP, Hua D, Desaire H. Glycosylation and disulfide bond analysis of transiently and stably expressed clade C HIV-1 gp140 trimers in 293T cells identifies disulfide heterogeneity present in both proteins and differences in O-linked glycosylation. J Proteome Res. 2014;13(9):4012–27.
Moulaei T, Stuchlik O, Reed M, Yuan W, Pohl J, Lu W, et al. Topology of the disulfide bonds in the antiviral lectin scytovirin. Protein Sci. 2010;19(9):1649–61.
Pike GM, Madden BJ, Melder DC, Charlesworth MC, Federspiel MJ. Simple, automated, high resolution mass spectrometry method to determine the disulfide bond and glycosylation patterns of a complex protein. J Biol Chem. 2011;286(20):17954–67.
Go EP, Zhang Z, Menon S, Desaire H. Analysis of the disulfide bond arrangement of the HIV envelope protein CON-S gp140 ΔCFI shows variability in the V1 and V2 regions. J Proteome Res. 2011;10(2):578–91.
Cramer CN, Kelstrup CD, Olsen JV, Haselmann KF, Nielsen PK. Complete mapping of complex disulfide patterns and closely-spaced cysteines by in-source reduction and data-dependent mass spectrometry. Anal Chem. 2017;89(11):5949–57.
Clark DF, Go EP, Toumi ML, Desaire H. Collision induced dissociation products of disulfide-bonded peptides: ions result from the cleavage of more than one bond. J Am Soc Mass Spectrom. 2011;22(3):492–8.
Wu S-L, Jiang H, Hancock W, Karger BL. Identification of the unpaired cysteine status and complete mapping of the 17 disulfides of recombinant tissue plasminogen activator using LC-MS with electron transfer dissociation/collision induced dissociation. Anal Chem. 2010;82:5296–303.
Choi S, Jeong J, Na S, Lee HS, Kim H-Y, Lee K-J, et al. New algorithm for the identification of intact disulfide linkages based on fragmentation characteristics in tandem mass spectra. J Proteome Res. 2010;9(1):626–35.
Chrisman PA, Pitteri SJ, Hogan JM, McLuckey SA. SO2 -* electron transfer ion/ion reactions with disulfide linked polypeptide ions. J Am Soc Mass Spectrom. 2005;16(7):1020–30.
Gunawardena HP, Gorenstein L, Erickson DE, Xia Y, McLuckey SA. Electron transfer dissociation of multiply protonated and fixed charge disulfide linked polypeptides. Int J Mass Spectrom. 2007;265(2-3):130–8.
Olsen JV, Macek B, Lange O, Makarov A, Horning S, Mann M. Higher-energy C-trap dissociation for peptide modification analysis. Nat Methods. 2007;4(9):709–12.
Frese CK, Altelaar AFM, van den Toorn H, Nolting D, Griep-Raming J, Heck AJR, et al. Toward full peptide sequence coverage by dual fragmentation combining electron-transfer and higher-energy collision dissociation tandem mass apectrometry. Anal Chem. 2012;84(22):9668–73.
Brunner AM, Lössl P, Liu F, Huguet R, Mullen C, Yamashita M, et al. Benchmarking multiple fragmentation methods on an Orbitrap fusion for top-down phospho-proteoform characterization. Anal Chem. 2015;87(8):4152–8.
Frese CK, Altelaar AFM, Hennrich ML, Nolting D, Zeller M, Griep-Raming J, et al. Improved peptide identification by targeted fragmentation using CID, HCD and ETC on an LTQ-Orbitrap Velos. J Proteome Res. 2011;10(5):2377–88.
Fung YME, Kjeldsen F, Silivra OA, Chan TWD, Zubarev RA. Facile disulfide bond cleavage in gaseous peptide and protein cations by ultraviolet photodissociation at 157 nm. Angew Chem Int Ed. 2005;44:6399–403.
Agarwal A, Diedrich JK, Julian RR. Direct elucidation of disulfide bond partners using ultraviolet photodissociation mass spectrometry. Anal Chem. 2011;83(17):6455–8.
Cole SR, Ma X, Zhang X, Xia Y. Electron transfer dissociation (ETD) of peptides containing intrachain disulfide bonds. J Am Soc Mass Spectrom. 2012;23(2):310–20.
Durand KL, Tan L, Stinson CA, Love-Nkansah CB, Ma X, Xia Y. Assigning peptide disulfide linkage pattern among regio-isomers via methoxy addition to disulfide and tandem mass spectrometry. J Am Soc Mass Spectrom. 2017;28(6):1099–108.
Massonnet P, Upert G, Smargiasso N, Gilles N, Quinton L, De Pauw E. Combined use of ion mobility and collision-induced dissociation to investigate the opening of disulfide bridges by electron-transfer dissociation in peptides bearing two disulfide bonds. Anal Chem. 2015;87(10):5240–6.
Mikesh LM, Ueberheide B, Chi A, Coon JJ, Syka JEP, Shabanowitz J, et al. The utility of ETD mass spectrometry in proteomic analysis. Biochim Biophys Acta. 2006;1764(12):1811–22.
Lu C, Liu D, Liu H, Motchnik P. Characterization of monoclonal antibody size variants containing extra light chains. mAbs. 2013;5(1):102–13.
Klapoetke S, Xie MH. Disulfide bond characterization of human factor Xa by mass spectrometry through protein-level partial reduction. J Pharmaceut Biomed. 2017;132:238–46.
Albert A, Eksteen JI, Isaksson J, Sengee M, Hansen T, Vasskog T. General approach to determine disulfide connectivity in cysteine-rich peptides by sequential alkylation on solid phase and mass spectrometry. Anal Chem. 2016;88:9539–46.
Li X, Xu W, Paporello B, Richardson D, Liu H. Liquid chromatography and mass spectrometry with post-column partial reduction for the analysis of native and scrambled disulfide bonds. Anal Biochem. 2013;439(2):184–6.
Li X, Wang F, Xu W, May K, Richardson D, Liu H. Disulfide bond assignment of an IgG1 monoclonal antibody by LC-MS with post-column partial reduction. Anal Biochem. 2013;436(2):93–100.
Liu H, Lei QP, Washabaugh M. Characterization of IgG2 disulfide bonds with LC/MS/MS and postcolumn online reduction. Anal Chem. 2016;88(10):5080–7.
Cramer CN, Haselmann KF, Olsen JV, Nielsen PK. Disulfide linkage characterization of disulfide bond-containing proteins and peptides by reducing electrochemistry and mass spectrometry. Anal Chem. 2016;88(3):1585–92.
Switzar L, Nicolardi S, Rutten JW, Oberstein SA, Aartsma-Rus A, van der Burgt YE. In-depth characterization of protein disulfide bonds by online liquid chromatography-electrochemistry-mass spectrometry. J Am Soc Mass Spectrom. 2016;27(1):50–8.
Zheng Q, Zhang H, Chen H. Integration of online digestion and electrolytic reduction with mass spectrometry for rapid disulfide-containing protein structural analysis. Int J Mass Spectrom. 2013;353:84–92.
Zhang Y, Dewald HD, Chen H. Online mass spectrometric analysis of proteins/peptides following electrolytic cleavage of disulfide bonds. J Proteome Res. 2011;10(3):1293–304.
Jiang H, Wu S-L, Karger BL, Hancock WS. Mass spectrometric analysis of innovator, counterfeit, and follow-on recombinant human growth hormone. Biotechnol Prog. 2009;25(1):207–18.
Xiang T, Chumsae C, Liu H. Localization and quantitation of free sulfhydryl in recombinant monoclonal antibodies by differential labeling with 12C and 13C iodoacetic acid and LC-MS analysis. Anal Chem. 2009;81(19):8101–8.
Chumsae C, Gaza-Bulseco G, Liu H. Identification and localization of unpaired cysteine residues in monoclonal antibodies by fluorescence labeling and mass spectrometry. Anal Chem. 2009;81(15):6449–57.
Xu H, Zhang L, Freitas MA. Identification and characterization of disulfide bonds in proteins and peptides from tandem MS data by use of the MassMatrix and MS/MS search engine. J Proteome Res. 2008;7(1):138–44.
Murad W, Singh R, Yen TY. An efficient algorithmic approach for mass spectrometry-based disulfide connectivity determination using multi-ion analysis. BMC Bioinformatics. 2011;12(Suppl 1):S12.
Murad W, Singh R. MS2DB+: a software for determination of disulfide bonds using multi-ion analysis. IEEE Trans Nanobioscience. 2013;12(2):69–71.
Huang SY, Wen CH, Li DT, Hsu JL, Chen C, Shi FK, et al. Assignment of disulfide-linked peptides using automatic a1 ion recognition. Anal Chem. 2008;80(23):9135–40.
Huang S-Y, Hsieh Y-T, Chen C-H, Chen C-C, Sung W-C, Chou M-Y, et al. Automatic disulfide bond assignment using a1 ion screening by mass spectrometry for structural characterization of protein pharmaceuticals. Anal Chem. 2012;84(11):4900–6.
Huang SY, Chen SF, Chen CH, Huang HW, Wu WG, Sung WC. Global disulfide bond profiling for crude snake venom using dimethyl labeling coupled with mass spectrometry and RADAR algorithm. Anal Chem. 2014;86(17):8742–50.
Goyder MS, Rebeaud F, Pfeifer ME, Kalman F. Strategies in mass spectrometry for the assignment of Cys-Cys disulfide connectivities in proteins. Expert Rev Proteomics. 2013;10(5):489–501.
Bagal D, Valliere-Douglass JF, Balland A, Schnier PD. Resolving disulfide structural isoforms of IgG2 monoclonal antibodies by ion mobility mass spectrometry. Anal Chem. 2010;82(16):6751–5.
Wang Q, Lacher NA, Muralidhara BK, Schlittler MR, Aykent S, Demarest CW. Rapid and refined separation of human IgG2 disulfide isomers using superficially porous particles. J Sep Sci. 2010;33(17-18):2671–80.
Cao X, He Y, Smith J, Wirth MJ. Alleviating nonlinear behavior of disulfide isoforms in the reversed-phase liquid chromatography of IgG2. J Chromatogr A. 2015;1410:147–53.
Acknowledgements
The authors acknowledge funding from the National Institutes of Health (grants R01AI125093 and R01AI094797) provided to HD.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Rights and permissions
About this article
Cite this article
Lakbub, J.C., Shipman, J.T. & Desaire, H. Recent mass spectrometry-based techniques and considerations for disulfide bond characterization in proteins. Anal Bioanal Chem 410, 2467–2484 (2018). https://doi.org/10.1007/s00216-017-0772-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00216-017-0772-1