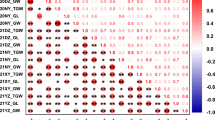
Abstract
Genome-wide association study (GWAS) has become an obvious general approach for studying traits of agricultural importance in higher plants, especially crops. Here, we present a GWAS of 32 morphologic and 10 agronomic traits in a collection of 615 barley cultivars genotyped by genome-wide polymorphisms from a recently developed barley oligonucleotide pool assay. Strong population structure effect related to mixed sampling based on seasonal growth habit and ear row number is present in this barley collection. Comparison of seven statistical approaches in a genome-wide scan for significant associations with or without correction for confounding by population structure, revealed that in reducing false positive rates while maintaining statistical power, a mixed linear model solution outperforms genomic control, structured association, stepwise regression control and principal components adjustment. The present study reports significant associations for sixteen morphologic and nine agronomic traits and demonstrates the power and feasibility of applying GWAS to explore complex traits in highly structured plant samples.



Similar content being viewed by others
References
Alonso-Blanco C, Aarts MGM, Bentsink L, Keurentjes JJB, Reymond M, Vreugdenhil D, Koornneef M (2009) What has natural variation taught us about plant development, physiology, and adaptation? Plant Cell 21:1877–1896
Astle W, Balding DJ (2009) Population structure and cryptic relatedness in genetic association studies. Stat Sci 24:451–471
Atwell S, Huang YS, Vilhjalmsson BJ, Willems G, Horton M, Li Y, Meng D, Platt A, Tarone AM, Hu TT, Jiang R, Muliyati NW, Zhang X, Amer MA, Baxter I, Brachi B, Chory J, Dean C, Debieu M, de Meaux J, Ecker JR, Faure N, Kniskern JM, Jones JDG, Michael T, Nemri A, Roux F, Salt DE, Tang C, Todesco M, Traw MB, Weigel D, Marjoram P, Borevitz JO, Bergelson J, Nordborg M (2010) Genome-wide association study of 107 traits in Arabidopsis thaliana inbred lines. Nature 465:627–631
Aulchenko YS, de Koning D-J, Haley C (2007) Genomewide rapid association using mixed model and regression: a fast and simple method for genomewide pedigree-based quantitative trait loci association analysis. Genetics 177:577–585
Bacanu SA, Devlin B, Roeder K (2002) Association studies for quantitative traits in structured populations. Genet Epidemiol 22:78–93
Balding D (2006) A tutorial on statistical methods for population association studies. Nat Rev Genet 7:781–791
Bennetzen JL, Ma J (2003) The genetic colinearity of rice and other cereals on the basis of genomic sequence analysis. Curr Opin Plant Biol 6:128–133
Bewley JD (1997) Seed germination and dormancy. Plant Cell 9:1055–1066
Close T, Bhat P, Lonardi S, Wu Y, Rostoks N, Ramsay L, Druka A, Stein N, Svensson J, Wanamaker S, Bozdag S, Roose M, Moscou M, Chao S, Varshney R, Szucs P, Sato K, Hayes P, Matthews D, Kleinhofs A, Muehlbauer G, DeYoung J, Marshall D, Madishetty K, Fenton R, Condamine P, Graner A, Waugh R (2009) Development and implementation of high-throughput SNP genotyping in barley. BMC Genom 10:582
Cockram J, White J, Leigh FJ, Lea VJ, Chiapparino E, Laurie DA, Mackay IJ, Powell W, O’Sullivan DM (2008) Association mapping of partitioning loci in barley. BMC Genet 9:16
Cockram J, White J, Zuluaga DL, Smith D, Comadran J, Macaulay M, Luo Z, Kearsey MJ, Werner P, Harrap D, Tapsell C, Liu H, Hedley PE, Stein N, Schulte D, Steuernagel B, Marshall DF, Thomas WTB, Ramsay L, Mackay I, Balding DJ, Consortium TA, Waugh R, O’Sullivan DM (2010) Genome-wide association mapping to candidate polymorphism resolution in the unsequenced barley genome. Proc Natl Acad Sci USA (published ahead of print)
Devlin B, Roeder K (1999) Genomic control for association studies. Biometrics 55:997–1004
Dubcovsky J, Ramakrishna W, SanMiguel PJ, Busso CS, Yan L, Shiloff BA, Bennetzen JL (2001) Comparative sequence analysis of colinear barley and rice bacterial artificial chromosomes. Plant Physiol 125:1342–1353
Falconer DS, Mackay TFC (1996) Introduction to quantitative genetics, 4th edition. Longman Press, New York
Goyal K, Walton LJ, Tunnacliffe A (2005) LEA proteins prevent protein aggregation due to water stress. Biochem J 388:151–157
Gupta PK, Rustgi S, Kulwal PL (2005) Linkage disequilibrium and association studies in higher plants: present status and future prospects. Plant Mol Biol 57:461–485
Hamblin MT, Close TJ, Bhat PR, Chao S, Kling JG, Abraham KJ, Blake T, Brooks WS, Cooper B, Griffey CA, Hayes PM, Hole DJ, Horsley RD, Obert DE, Smith KP, Ullrich SE, Muehlbauer GJ, Jannink J-L (2010) Population structure and linkage disequilibrium in U.S. Barley Germplasm: implications for association mapping. Crop Sci 50:556–566
Hayes PM, Castro A, Marquez-Cedillo L, Corey A, Henson C, Jones BL, Kling J, Mather D, Matus I, Rossi C, Sato K (2003) Genetic diversity for quantitatively inherited agronomic and malting quality traits. Diversity in Barley (Hordeum vulgare). Elsevier, Amsterdam
Hoggart CJ, Parra EJ, Shriver MD, Bonilla C, Kittles RA, Clayton DG, McKeigue PM (2003) Control of confounding of genetic associations in stratified populations. Am J Hum Genet 72:1492–1504
Kang HM, Zaitlen NA, Wade CM, Kirby A, Heckerman D, Daly MJ, Eskin E (2008) Efficient control of population structure in model organism association mapping. Genetics 178:1709–1723
Kang HM, Sul JH, Service SK, Zaitlen NA, Kong S-y, Freimer NB, Sabatti C, Eskin E (2010) Variance component model to account for sample structure in genome-wide association studies. Nat Genet 42:348–354
Komatsuda T, Pourkheirandish M, He C, Azhaguvel P, Kanamori H, Perovic D, Stein N, Graner A, Wicker T, Tagiri A, Lundqvist U, Fujimura T, Matsuoka M, Matsumoto T, Yano M (2007) Six-rowed barley originated from a mutation in a homeodomain-leucine zipper I-class homeobox gene. Proc Natl Acad Sci USA 104:1424–1429
Kraakman ATW, Niks RE, Van den Berg PMMM, Stam P, Van Eeuwijk FA (2004) Linkage disequilibrium mapping of yield and yield stability in modern spring barley cultivars. Genetics 168:435–446
Mackay I, Powell W (2007) Methods for linkage disequilibrium mapping in crops. Trends Plant Sci 12:57–63
Malysheva-Otto LV, Ganal MW, Roder MS (2006) Analysis of molecular diversity, population structure and linkage disequilibrium in a worldwide survey of cultivated barley germplasm (Hordeum vulgare L.). BMC Genet 7:6
Moose SP, Mumm RH (2008) Molecular plant breeding as the foundation for 21st century crop improvement. Plant Physiol 147:969–977
Pelleg D, Moore A (2000) X-means: extending k-means with efficient estimation of the number of clusters. In: Proceedings of the seventeenth international conference on machine learning. Morgan Kaufmann, San Francisco, pp 727–734
Platt A, Horton M, Huang YS, Li Y, Anastasio AE, Mulyati NW, Ågren J, Bossdorf O, Byers D, Donohue K, Dunning M, Holub EB, Hudson A, Le Corre V, Loudet O, Roux F, Warthmann N, Weigel D, Rivero L, Scholl R, Nordborg M, Bergelson J, Borevitz JO (2010) The scale of population structure in Arabidopsis thaliana. PLoS Genet 6:e1000843
Potokina E, Druka A, Luo Z, Wise R, Waugh R, Kearsey M (2008) Gene expression quantitative trait locus analysis of 16000 barley genes reveals a complex pattern of genome-wide transcriptional regulation. Plant J 53:90–101
Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D (2006) Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet 38:904–909
Price AL, Zaitlen NA, Reich D, Patterson N (2010) New approaches to population stratification in genome-wide association studies. Nat Rev Genet 11:459–463
Pritchard JK, Stephens M, Donnelly P (2000a) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Pritchard JK, Stephens M, Rosenberg NA, Donnelly P (2000b) Association mapping in structured populations. Am J Hum Genet 67:170–181
Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MAR, Bender D, Maller J, Sklar P, de Bakker PIW, Daly MJ, Sham PC (2007) PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 81:559–575
Remington DL, Thornsberry JM, Matsuoka Y, Wilson LM, Whitt SR, Doeblay J, Kresovich S, Goodman MM, Buckler ES (2001) Structure of linkage disequilibrium and phenotypic associations in the maize genome. Proc Natl Acad Sci USA 98:11479–11484
Rostoks N, Ramsay L, MacKenzie K, Cardle L, Bhat PR, Roose ML, Svensson JT, Stein N, Varshney RK, Marshall DF, Grainer A, Close TJ, Waugh R (2006) Recent history of artificial outcrossing facilitates whole-genome association mapping in elite inbred crop varieties. Proc Natl Acad Sci USA 103:18656–18661
Schmalenbach I, Pillen K (2009) Detection and verification of malting quality QTLs using wild barley introgression lines. Theor Appl Genet 118:1411–1427
Setakis E, Stirnadel H, Balding DJ (2006) Logistic regression protects against population structure in genetic association studies. Genome Res 16:290–296
Simon-Sanchez J, Schulte C, Bras JM, Sharma M, Gibbs JR, Berg D, Paisan-Ruiz C, Lichtner P, Scholz SW, Hernandez DG, Kruger R, Federoff M, Klein C, Goate A, Perlmutter J, Bonin M, Nalls MA, Illig T, Gieger C, Houlden H, Steffens M, Okun MS, Racette BA, Cookson MR, Foote KD, Fernandez HH, Traynor BJ, Schreiber S, Arepalli S, Zonozi R, Gwinn K, van der Brug M, Lopez G, Chanock SJ, Schatzkin A, Park Y, Hollenbeck A, Gao JJ, Huang XM, Wood NW, Lorenz D, Deuschl G, Chen HL, Riess O, Hardy JA, Singleton AB, Gasser T (2009) Genome-wide association study reveals genetic risk underlying Parkinson’s disease. Nat Genet 41:1308–1309
Sneller CH, Mather DE, Crepieux S (2009) Analytical approaches and population types for finding and utilizing QTL in complex plant populations. Crop Sci 49:363–380
Sun G, Zhu C, Kramer MH, Yang SS, Song W, Piepho HP, Yu J (2010) Variation explained in mixed-model association mapping. Heredity 105:333–340
Szűcs P, Skinner J, Karsai I, Cuesta-Marcos A, Haggard K, Corey A, Chen T, Hayes P (2007) Validation of the VRN-H2/VRN-H1 epistatic model in barley reveals that intron length variation in VRN-H1 may account for a continuum of vernalization sensitivity. Mol Genet Genom 277:249–261
Taketa S, Amano S, Tsujino Y, Sato T, Saisho D, Kakeda K, Nomura M, Suzuki T, Matsumoto T, Sato K, Kanamori H, Kawasaki S, Takeda K (2008) Barley grain with adhering hulls is controlled by an ERF family transcription factor gene regulating a lipid biosynthesis pathway. Proc Natl Acad Sci USA 105:4062–4067
von Zitzewitz J, Szucs P, Dubcovsky J, Yan L, Francia E, Pecchioni N, Casas A, Chen TH, Hayes PM, Skinner JS (2005) Molecular and structural characterization of barley vernalization genes. Plant Mol Biol 59:449–467
Wilk MB, Gnanadesikan R (1968) Probability plotting methods for the analysis of data. Biometrika 55:1–17
Wu C, DeWan A, Hoh J, Wang Z (2010) A comparison of association methods correcting for population stratification in case–control studies. Ann Hum Genet 00:1–10
Yan L, Fu D, Li C, Blechl A, Tranquilli G, Bonafede M, Sanchez A, Valarik M, Yasuda S, Dubcovsky J (2006) The wheat and barley vernalization gene VRN3 is an orthologue of FT. Proc Natl Acad Sci USA 103:19581–19586
Yu JM, Pressoir G, Briggs WH, Bi IV, Yamasaki M, Doebley JF, McMullen MD, Gaut BS, Nielsen DM, Holland JB, Kresovich S, Buckler ES (2006) A unified mixed-model method for association mapping that accounts for multiple levels of relatedness. Nat Genet 38:203–208
Zhao K, Aranzana MJ, Kim S, Lister C, Shindo C, Tang C, Toomajian C, Zheng H, Dean C, Marjoram P, Nordborg M (2007) An Arabidopsis example of association mapping in structured samples. PLoS Genet 3:e4
Ziv E, Burchard EG (2003) Human population structure and genetic association studies. Pharmacogenomics 4:431–441
Acknowledgments
This study was supported by research grants for the ‘Association Genetics of UK Elite Barley’ project, which was funded by BBSRC and RERAD as part of the Sustainable Arable LINK programme with industrial support from HGCA, KWS (UK), LS Plant Breeding, Syngenta Seeds, Groupe Limagrain, Secobra UK, Svalof Weibull, Perten Instruments AB, The Maltsters Association of Great Britain, The Scotch Whisky Research Institute and Campden BRi. ZWL is also supported by the Leverhulme Trust (RCEJ1471) of UK, NSFC (31071084) and The Basic Research Program (2012CB316505) of China.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by J. Yu.
An erratum to this article can be found at http://dx.doi.org/10.1007/s00122-011-1771-9
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Wang, M., Jiang, N., Jia, T. et al. Genome-wide association mapping of agronomic and morphologic traits in highly structured populations of barley cultivars. Theor Appl Genet 124, 233–246 (2012). https://doi.org/10.1007/s00122-011-1697-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-011-1697-2