Abstract
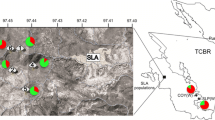
Genetic diversity and relatedness were studied in 30 Norwegian local populations of meadow fescue (Festuca pratensis Huds.) using amplified fragment length polymorphism (AFLP) markers. The populations were also compared with 13 Nordic meadow fescue cultivars in order to analyse the distribution of variation in local populations and cultivars and to elucidate relationships between local populations and cultivars. Analysis of molecular variance (amova) analysis showed that most of the variation was present within populations and that little variation was found between local populations and cultivars. Separate amova analyses of local populations and cultivars revealed a higher level of variation within registered cultivars than within local populations. A cluster analysis based on corrected average pairwise differences between populations showed that the populations could be divided into three clusters, of which one also contained the cultivars. These results were supported by principal coordinates analysis. The results indicate that the Norwegian meadow fescue has a narrow genetic basis and that the local populations in Norway can be divided into three groups following the most probable routes of introduction of the species into Norway. The inland populations are closely related to the cultivars and have most probably been established as a result of migration from sown meadows. The western and southern populations probably originate from human activity—for example, trade—to the coastal western and northern parts of the country and to the central parts of southern Norway.




Similar content being viewed by others
References
Alm V, Fang C, Busso CS, Devos KM, Vollan K, Grieg Z, Rognli OA (2003) A linkage map of meadow fescue (Festuca pratensis Huds.) and comparative mapping with other Poaceae species. Theor Appl Genet 108:25–40
Balfourier F, Charmet G, Ravel C (1998) Genetic differentiation within and between natural populations of perennial and annual ryegrass (Lolium perenne and L. rigidum). Heredity 81:100–110
Blytt MN (1861) Norges flora, eller beskrivelser over de I Norge vildtvoxende karplanter: tilligemed angivelser af de geographiske forholde, under hvilke de forekomme: 1ste deel. Kongelige Norske Videnskabers Selskab, Christiania
Chowdhury MA, Slinkard AE (2000) Genetic diversity in grasspea (Lathyrus sativus L.). Genet Resour Crop Evol 47:163–169
Cresswell A, Sackville Hamilton NR, Roy AK, Viegas MF (2001) Use of amplified length polymorphism markers to assess genetic diversity of Lolium species from Portugal. Mol Ecol 10:229–241
Donini P, Elias ML, Bougourd SM, Koebner RMD (1997) AFLP fingerprinting reveals pattern differences between template DNA extracted from different plant organs. Genome 40:521–526
Elven R (1994) Lid & Lids Norsk flora, 6th edn. Det Norske Samlaget, Oslo, Norway
Excoffier L, Smouse PE, Quattro JM (1992) Analysis of molecular variance inferred from metric distances among DNA haplotypes—application to human mitochondrial-DNA restriction data. Genetics 131:479–491
Fægri K (1960) Maps of distribution of Norwegian plants 1. The coast plants. Skrifter Universitetet I Bergen 26, Oslo University Press, Oslo
Fjellheim S, Rognli OA (2005) Genetic diversity within and among Nordic meadow fescue (Festuca pratensis Huds.) cultivars based on AFLP markers. Crop Sci (in press)
Glærum O (1914) Engdyrkning og enkulturforsøk. In: Ødegaard N, Vik K, Klokk O (eds) Norsk Forsøksarbeid I Jordbruket. Grøndahl & Søns Forlag, Kristiania
Grivet D, Petit RJ (2003) Chloroplast DNA phylogeography of the hornbeam in Europe: evidence for a bottleneck at the outset of postglacial colonization. Conserv Genet 4:47–56
Guthridge KM, Dupal MP, Kölliker R, Jones ES, Smith KF, Forster JW (2001) AFLP analysis of genetic diversity within and between populations of perennial ryegrass (Lolium perenne L.). Euphytica 122:191–201
Hasund S (1925) Norges Landbruk 1875–1925. In: Lantbruket I Norden 1875–1925. Göteborgs Litografiska Aktiebolag, Göteborg, Sverige, pp 329–449
Hepburn AG, Belanger FC, Mattheis JR (1987) DNA methylation in plants. Dev Genet 8:475–493
Humphreys M, Humphreys J, Donnison I, King I, Thomas HM, Ghesquiere M, Durand JL, Rognli OA, Zwierzykowski Z, Rapacz M (2004) Molecular breeding and functional genomics for tolerance to abiotic stress. In: Hopkins A, Wang ZY, Mian R, Sledge M, Barker RE (eds) Molecular breeding of forage and turf. Dev Plant Breed 11:61–80
Karp A, Seberg O, Buiatti M (1996) Molecular techniques in the assessment of botanical diversity. Ann Bot 78:143–149
Kölliker R, Stadelmann FJ, Reidy B, Nosberger J (1998) Fertilization and defoliation frequency affect genetic diversity of Festuca pratensis Huds. in permanent grasslands. Mol Ecol 7:1557–1567
Lundqvist A (1962) The nature of the two-loci incompatibility system in grasses. II. Number of alleles at the incompatibility loci in Festuca pratensis Huds. Hereditas 48:169–181
Lynch M, Milligan BG (1994) Analysis of population genetic structure with RAPD markers. Mol Ecol 3:91–99
Nei M (1987) Molecular evolutionary genetics. Columbia University Press, New York, NY, USA
Nybom H (2004) Comparison of different nuclear DNA markers for estimating intraspecific genetic diversity in plants. Mol Ecol 13:1143–1155
Rognli OA, Nilsson NO, Nurminiemi M (2000) Effects of distance and pollen competition on gene flow in the wind-pollinated grass Festuca pratensis Huds. Heredity 85:550–560
Rohlf FJ (2000) ntsys-pc. Numerical taxonomy and multivariate analysis system, version 2.1. Exeter Software, New York, USA
Schneider S, Roessli D, Excoffier L (2000) arlequin ver. 2.0: a software for population genetics data analysis. Genetics and Biometry Laboratory, University of Geneva, Switzerland
Sharp PJ, Kreis M, Shewry PR, Gale MD (1988) Location of beta amylase sequences in wheat and its relatives. Theor Appl Genet 75:286–290
Stewart CN, Excoffier L (1996) Assessing population genetic structure and variability with RAPD data: Application to Vaccinium macrocarpon (American Cranberry). J Evol Biol 9:153–171
Tajima F (1983) Evolutionary relationship of DNA sequences in finite populations. Genetics 105:437–460
Tajima F (1993) Measurement of DNA polymorphism. In: Takahata N, Clark AG (eds) Mechanisms of molecular evolution. Introduction to molecular paleopopulation biology. Sinauer Associates, Sunderland, pp 37–59
Theiss G, Follmann H (1980) 5-Methylcytosine formation in wheat embryo DNA. Biochem Biophys Res Commun 94:291–297
Vavilov NI (1940) The new systematics of cultivated plants. In: Huxley J (ed) The new systematics. Clarendon press, Oxford, pp 549–566
Vos P, Hogers R, Bleeker M, Reijans M, Vandelee T, Hornes M, Frijters A, Pot J, Peleman J, Kuiper M, Zabeau M (1995) AFLP—a new technique for DNA-fingerprinting. Nucleic Acids Res 23:4407–4414
Yamada T, Momotaz A (2004) Usefulness of simple sequence repeat (SSR) polymorphism for the breeding programs in Festulolium. In: Yamada T, Takamizo T (eds) Development of a novel grass with environmental stress tolerance and high forage quality through intergeneric hybridization between Lolium and Festuca. National Agriculture and Bio-oriented Research Organization, pp 43–48
Zhivotovsky LA (1999) Estimating population structure in diploids with multilocus dominant DNA markers. Mol Ecol 8:907–913
Acknowledgements
This investigation was supported by a grant from the NGB (project no. AG4 19). The authors thank Zanina Grieg, Vibeke Alm and Ingvild Marum for their excellent technical assistance, Anders Bryn for help with creating the sample map and Robert Koebner, Siri Grønnerød and Reidar Elven for their helpful comments on the manuscript.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by T. Lübberstedt
Rights and permissions
About this article
Cite this article
Fjellheim, S., Rognli, O.A. Molecular diversity of local Norwegian meadow fescue (Festuca pratensis Huds.) populations and Nordic cultivars—consequences for management and utilisation. Theor Appl Genet 111, 640–650 (2005). https://doi.org/10.1007/s00122-005-2006-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-005-2006-8